Loci Encoding Compounds Potentially Active against Drug-Resistant Pathogens amidst a Decreasing Pool of Novel Antibiotics
- PMID: 31540982
- PMCID: PMC6856318
- DOI: 10.1128/AEM.01438-19
Loci Encoding Compounds Potentially Active against Drug-Resistant Pathogens amidst a Decreasing Pool of Novel Antibiotics
Abstract
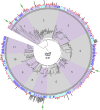
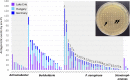
Since the discovery of penicillin, microbes have been a source of antibiotics that inhibit the growth of pathogens. However, with the evolution of multidrug-resistant (MDR) strains, it remains unclear if there is an abundant or limited supply of natural products to be discovered that are effective against MDR isolates. To identify strains that are antagonistic to pathogens, we examined a set of 471 globally derived environmental Pseudomonas strains (env-Ps) for activity against a panel of 65 pathogens including Achromobacter spp., Burkholderia spp., Pseudomonas aeruginosa, and Stenotrophomonas spp. isolated from the lungs of cystic fibrosis (CF) patients. From more than 30,000 competitive interactions, 1,530 individual inhibitory events were observed. While strains from water habitats were not proportionate in antagonistic activity, MDR CF-derived pathogens (CF-Ps) were less susceptible to inhibition by env-Ps, suggesting that fewer natural products are effective against MDR strains. These results advocate for a directed strategy to identify unique drugs. To facilitate discovery of antibiotics against the most resistant pathogens, we developed a workflow in which phylogenetic and antagonistic data were merged to identify strains that inhibit MDR CF-Ps and subjected those env-Ps to transposon mutagenesis. Six different biosynthetic gene clusters (BGCs) were identified from four strains whose products inhibited pathogens including carbapenem-resistant P. aeruginosa BGCs were rare in databases, suggesting the production of novel antibiotics. This strategy can be utilized to facilitate the discovery of needed antibiotics that are potentially active against the most drug-resistant pathogens.IMPORTANCE Carbapenem-resistant P. aeruginosa is difficult to treat and has been deemed by the World Health Organization as a priority one pathogen for which antibiotics are most urgently needed. Although metagenomics and bioinformatic studies suggest that natural bacteria remain a source of novel compounds, the identification of genes and their products specific to activity against MDR pathogens remains problematic. Here, we examine water-derived pseudomonads and identify gene clusters whose compounds inhibit CF-derived MDR pathogens, including carbapenem-resistant P. aeruginosa.
Keywords: Pseudomonas; antagonistic; antibiotic; biosynthetic gene cluster; multidrug resistance; transposon mutagenesis.
Copyright © 2019 Basalla et al.
Figures



Similar articles
-
Environmental Pseudomonads Inhibit Cystic Fibrosis Patient-Derived Pseudomonas aeruginosa.Appl Environ Microbiol. 2016 Dec 30;83(2):e02701-16. doi: 10.1128/AEM.02701-16. Print 2017 Jan 15. Appl Environ Microbiol. 2016. PMID: 27881418 Free PMC article.
-
Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli.Drugs. 2019 Feb;79(3):271-289. doi: 10.1007/s40265-019-1055-2. Drugs. 2019. PMID: 30712199 Review.
-
The Inhibition Effect of Lactobacilli Against Growth and Biofilm Formation of Pseudomonas aeruginosa.Probiotics Antimicrob Proteins. 2018 Mar;10(1):34-42. doi: 10.1007/s12602-017-9267-9. Probiotics Antimicrob Proteins. 2018. PMID: 28293865
-
In vitro antimicrobial activity of ceftolozane/tazobactam against Pseudomonas aeruginosa and other non-fermenting Gram-negative bacteria in adults with cystic fibrosis.J Glob Antimicrob Resist. 2018 Sep;14:224-227. doi: 10.1016/j.jgar.2018.03.002. Epub 2018 Mar 17. J Glob Antimicrob Resist. 2018. PMID: 29559421
-
Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis.Am J Respir Med. 2003;2(4):321-32. doi: 10.1007/BF03256660. Am J Respir Med. 2003. PMID: 14719998 Review.
Cited by
-
Tradeoffs Between Evolved Phage Resistance and Antibiotic Susceptibility in a Highly Drug-Resistant Cystic Fibrosis-Derived Pseudomonas aeruginosa Strain.Phage (New Rochelle). 2024 Jun 21;5(2):45-52. doi: 10.1089/phage.2023.0022. eCollection 2024 Jun. Phage (New Rochelle). 2024. PMID: 39119204 Free PMC article.
-
Genome Mining for Antimicrobial Compounds in Wild Marine Animals-Associated Enterococci.Mar Drugs. 2021 Jun 6;19(6):328. doi: 10.3390/md19060328. Mar Drugs. 2021. PMID: 34204046 Free PMC article.
-
Novel Lytic Phages Protect Cells and Mice against Pseudomonas aeruginosa Infection.J Virol. 2021 Mar 25;95(8):e01832-20. doi: 10.1128/JVI.01832-20. Epub 2021 Jan 20. J Virol. 2021. PMID: 33472935 Free PMC article.
-
Deep learning approaches for natural product discovery from plant endophytic microbiomes.Environ Microbiome. 2021 Mar 18;16:6. doi: 10.1186/s40793-021-00375-0. eCollection 2021. Environ Microbiome. 2021. PMID: 33758794 Free PMC article. Review.
-
Opportunistic pathogens and polycocktail drugs fuel dynamic public health threats during the opioid crisis.PLoS One. 2025 Aug 12;20(8):e0326200. doi: 10.1371/journal.pone.0326200. eCollection 2025. PLoS One. 2025. PMID: 40794717 Free PMC article.
References
-
- WHO. 27 February 2017. WHO publishes list of bacteria for which new antibiotics are urgently needed. World Health Organization, Geneva, Switzerland: https://www.who.int/en/news-room/detail/27-02-2017-who-publishes-list-of....
-
- O’Neill J. 2016. Tackling drug resistance infections globally: final report and recommendations: the review on antimicrobial resistance. Government of the United Kingdom, London, United Kingdom: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20c....
-
- Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi:10.1016/j.cell.2014.06.034. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

