T Cell-Specific Adaptor Protein Regulates Mitochondrial Function and CD4+ T Regulatory Cell Activity In Vivo following Transplantation
- PMID: 31541025
- PMCID: PMC6783373
- DOI: 10.4049/jimmunol.1801604
T Cell-Specific Adaptor Protein Regulates Mitochondrial Function and CD4+ T Regulatory Cell Activity In Vivo following Transplantation
Abstract
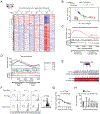
The T cell-specific adaptor protein (TSAd), encoded by the SH2D2A gene, is an intracellular molecule that binds Lck to elicit signals that result in cytokine production in CD4+ T effector cells (Teff). Nevertheless, using Sh2d2a knockout (KO; also called TSAd-/-) mice, we find that alloimmune CD4+ Teff responses are fully competent in vivo. Furthermore, and contrary to expectations, we find that allograft rejection is accelerated in KO recipients of MHC class II-mismatched B6.C-H-2bm12 heart transplants versus wild-type (WT) recipients. Also, KO recipients of fully MHC-mismatched cardiac allografts are resistant to the graft-prolonging effects of costimulatory blockade. Using adoptive transfer models, we find that KO T regulatory cells (Tregs) are less efficient in suppressing Teff function and they produce IFN-γ following mitogenic activation. In addition, pyrosequencing demonstrated higher levels of methylation of CpG regions within the Treg-specific demethylated region of KO versus WT Tregs, suggesting that TSAd, in part, promotes Treg stability. By Western blot, Lck is absent in the mitochondria of KO Tregs, and reactive oxygen species production by mitochondria is reduced in KO versus WT Tregs. Full transcriptomic analysis demonstrated that the key mechanism of TSAd function in Tregs relates to its effects on cellular activation rather than intrinsic effects on mitochondria/metabolism. Nevertheless, KO Tregs compensate for a lack of activation by increasing the number of mitochondria per cell. Thus, TSAd serves as a critical cell-intrinsic molecule in CD4+Foxp3+ Tregs to regulate the translocation of Lck to mitochondria, cellular activation responses, and the development of immunoregulation following solid organ transplantation.
Copyright © 2019 by The American Association of Immunologists, Inc.
Conflict of interest statement
Conflicts of Interest
Potential conflicts of interest have been reviewed by the Office of General Counsel at Boston Children’s Hospital. The authors of this manuscript declare no conflicts of interest.
Figures
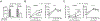


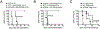

References
-
- Spurkland A,Brinchmann JE,Markussen G,Pedeutour F,Munthe E,Lea T,Vartdal F, and Aasheim HC. 1998. Molecular cloning of a T cell-specific adapter protein (TSAd) containing an Src homology (SH) 2 domain and putative SH3 and phosphotyrosine binding sites. J Biol Chem. 273: 4539–46. - PubMed
-
- Dai KZ,Vergnaud G,Ando A,Inoko H, and Spurkland A. 2000. The SH2D2A gene encoding the T-cell-specific adapter protein (TSAd) is localized centromeric to the CD1 gene cluster on human Chromosome 1. Immunogenetics. 51: 179–85. - PubMed
-
- Choi YB,Kim CK, and Yun Y. 1999. Lad, an adapter protein interacting with the SH2 domain of p56lck, is required for T cell activation. J Immunol. 163: 5242–9. - PubMed
-
- Lapinski PE,Oliver JA,Bodie JN,Marti F, and King PD. 2009. The T-cell-specific adapter protein family: TSAd, ALX, and SH2D4A/SH2D4B. Immunol Rev. 232: 240–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

