Time elapsed between Zika and dengue virus infections affects antibody and T cell responses
- PMID: 31541110
- PMCID: PMC6754404
- DOI: 10.1038/s41467-019-12295-2
Time elapsed between Zika and dengue virus infections affects antibody and T cell responses
Abstract
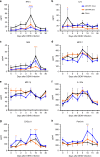
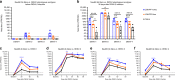
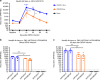
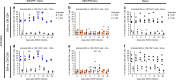
Zika virus (ZIKV) and dengue virus (DENV) are co-endemic in many parts of the world, but the impact of ZIKV infection on subsequent DENV infection is not well understood. Here we show in rhesus macaques that the time elapsed after ZIKV infection affects the immune response to DENV infection. We show that previous ZIKV exposure increases the magnitude of the antibody and T cell responses against DENV. The time interval between ZIKV and subsequent DENV infection further affects the immune response. A mid-convalescent period of 10 months after ZIKV infection results in higher and more durable antibody and T cell responses to DENV infection than a short period of 2 months. In contrast, previous ZIKV infection does not affect DENV viremia or pro-inflammatory status. Collectively, we find no evidence of a detrimental effect of ZIKV immunity in a subsequent DENV infection. This supports the implementation of ZIKV vaccines that could also boost immunity against future DENV epidemics.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

