TmDOTP: An NMR-based thermometer for magic angle spinning NMR experiments
- PMID: 31541931
- PMCID: PMC7296554
- DOI: 10.1016/j.jmr.2019.106574
TmDOTP: An NMR-based thermometer for magic angle spinning NMR experiments
Abstract
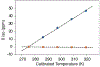
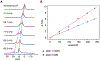
Solid state NMR is a powerful tool to probe membrane protein structure and dynamics in native lipid membranes. Sample heating during solid state NMR experiments can be caused by magic angle spinning and radio frequency irradiation such heating produces uncertainties in the sample temperature and temperature distribution, which can in turn lead to line broadening and sample deterioration. To measure sample temperatures in real time and to quantify thermal gradients and their dependence on radio frequency irradiation or spinning frequency, we use the chemical shift thermometer TmDOTP, a lanthanide complex. The H6 TmDOTP proton NMR peak has a large chemical shift (-176.3 ppm at 275 K) and it is well resolved from the protein and lipid proton spectrum. Compared to other NMR thermometers (e.g., the proton NMR signal of water), the proton spectrum of TmDOTP, particularly the H6 proton line, exhibits very high thermal sensitivity and resolution. In MAS studies of proteoliposomes we identify two populations of TmDOTP with differing temperatures and dependency on the radio frequency irradiation power. We interpret these populations as arising from the supernatant and the pellet, which is sedimented during sample spinning. In this study, we demonstrate that TmDOTP is an excellent internal standard for monitoring real-time temperatures of biopolymers without changing their properties or obscuring their spectra. Real time temperature calibration is expected to be important for the interpretation of dynamics and other properties of biopolymers.
Keywords: Dielectric loss; Magic-angle spinning; Nuclear magnetic resonance; Real-time NMR temperature measurement; Sample Heating; TmDOTP(5-).
Copyright © 2019. Published by Elsevier Inc.
Figures





Similar articles
-
Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review.
-
207Pb chemical shift thermometer at high temperature for magic angle spinning experiments.Solid State Nucl Magn Reson. 1999 Nov;15(2):119-23. doi: 10.1016/s0926-2040(99)00039-9. Solid State Nucl Magn Reson. 1999. PMID: 10670904
-
An NMR thermometer for cryogenic magic-angle spinning NMR: the spin-lattice relaxation of (127)I in cesium iodide.J Magn Reson. 2011 Oct;212(2):460-3. doi: 10.1016/j.jmr.2011.08.021. Epub 2011 Aug 17. J Magn Reson. 2011. PMID: 21906982
-
LnDOTA-d8 , a versatile chemical-shift thermometer for 2 H solid-state NMR.Magn Reson Chem. 2022 Oct;60(10):1005-1013. doi: 10.1002/mrc.5303. Epub 2022 Aug 17. Magn Reson Chem. 2022. PMID: 35938541
-
Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy: Advances in Methodology and Applications.Chem Rev. 2023 Feb 8;123(3):918-988. doi: 10.1021/acs.chemrev.2c00197. Epub 2022 Dec 21. Chem Rev. 2023. PMID: 36542732 Free PMC article. Review.
Cited by
-
Informing NMR experiments with molecular dynamics simulations to characterize the dominant activated state of the KcsA ion channel.J Chem Phys. 2021 Apr 28;154(16):165102. doi: 10.1063/5.0040649. J Chem Phys. 2021. PMID: 33940802 Free PMC article.
-
Recent developments in deuterium solid-state NMR for the detection of slow motions in proteins.Solid State Nucl Magn Reson. 2021 Feb;111:101710. doi: 10.1016/j.ssnmr.2020.101710. Epub 2021 Jan 7. Solid State Nucl Magn Reson. 2021. PMID: 33450712 Free PMC article. Review.
-
1H-Detected Biomolecular NMR under Fast Magic-Angle Spinning.Chem Rev. 2022 May 25;122(10):9943-10018. doi: 10.1021/acs.chemrev.1c00918. Epub 2022 May 10. Chem Rev. 2022. PMID: 35536915 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

