Extension distribution for DNA confined in a nanochannel near the Odijk regime
- PMID: 31542006
- PMCID: PMC7027588
- DOI: 10.1063/1.5121305
Extension distribution for DNA confined in a nanochannel near the Odijk regime
Abstract
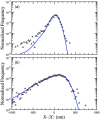
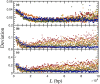
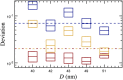
DNA confinement in a nanochannel typically is understood via mapping to the confinement of an equivalent neutral polymer by hard walls. This model has proven to be effective for confinement in relatively large channels where hairpin formation is frequent. An analysis of existing experimental data for Escherichia coli DNA extension in channels smaller than the persistence length, combined with an additional dataset for λ-DNA confined in a 34 nm wide channel, reveals a breakdown in this approach as the channel size approaches the Odijk regime of strong confinement. In particular, the predicted extension distribution obtained from the asymptotic solution to the weakly correlated telegraph model for a confined wormlike chain deviates significantly from the experimental distribution obtained for DNA confinement in the 34 nm channel, and the discrepancy cannot be resolved by treating the alignment fluctuations or the effective channel size as fitting parameters. We posit that the DNA-wall electrostatic interactions, which are sensible throughout a significant fraction of the channel cross section in the Odijk regime, are the source of the disagreement between theory and experiment. Dimensional analysis of the wormlike chain propagator in channel confinement reveals the importance of a dimensionless parameter, reflecting the magnitude of the DNA-wall electrostatic interactions relative to thermal energy, which has not been considered explicitly in the prevailing theories for DNA confinement in a nanochannel.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

