Ancestry-Dependent Enrichment of Deleterious Homozygotes in Runs of Homozygosity
- PMID: 31543216
- PMCID: PMC6817522
- DOI: 10.1016/j.ajhg.2019.08.011
Ancestry-Dependent Enrichment of Deleterious Homozygotes in Runs of Homozygosity
Abstract
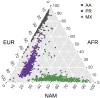
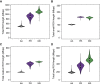
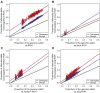
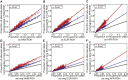
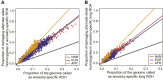
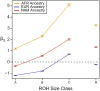
Runs of homozygosity (ROH) are important genomic features that manifest when an individual inherits two haplotypes that are identical by descent. Their length distributions are informative about population history, and their genomic locations are useful for mapping recessive loci contributing to both Mendelian and complex disease risk. We have previously shown that ROH, and especially long ROH that are likely the result of recent parental relatedness, are enriched for homozygous deleterious coding variation in a worldwide sample of outbred individuals. However, the distribution of ROH in admixed populations and their relationship to deleterious homozygous genotypes is understudied. Here we analyze whole-genome sequencing data from 1,441 unrelated individuals from self-identified African American, Puerto Rican, and Mexican American populations. These populations are three-way admixed between European, African, and Native American ancestries and provide an opportunity to study the distribution of deleterious alleles partitioned by local ancestry and ROH. We re-capitulate previous findings that long ROH are enriched for deleterious variation genome-wide. We then partition by local ancestry and show that deleterious homozygotes arise at a higher rate when ROH overlap African ancestry segments than when they overlap European or Native American ancestry segments of the genome. These results suggest that, while ROH on any haplotype background are associated with an inflation of deleterious homozygous variation, African haplotype backgrounds may play a particularly important role in the genetic architecture of complex diseases for admixed individuals, highlighting the need for further study of these populations.
Keywords: ROH; admixture; deleterious alleles; haplotype; homozygosity; identity by descent; population bottleneck; runs of homozygosity.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Long runs of homozygosity are enriched for deleterious variation.Am J Hum Genet. 2013 Jul 11;93(1):90-102. doi: 10.1016/j.ajhg.2013.05.003. Epub 2013 Jun 6. Am J Hum Genet. 2013. PMID: 23746547 Free PMC article.
-
Runs of homozygosity and distribution of functional variants in the cattle genome.BMC Genomics. 2015 Jul 22;16(1):542. doi: 10.1186/s12864-015-1715-x. BMC Genomics. 2015. PMID: 26198692 Free PMC article.
-
Fine-Scale Resolution of Runs of Homozygosity Reveal Patterns of Inbreeding and Substantial Overlap with Recessive Disease Genotypes in Domestic Dogs.G3 (Bethesda). 2019 Jan 9;9(1):117-123. doi: 10.1534/g3.118.200836. G3 (Bethesda). 2019. PMID: 30429214 Free PMC article.
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
-
Runs of homozygosity: windows into population history and trait architecture.Nat Rev Genet. 2018 Apr;19(4):220-234. doi: 10.1038/nrg.2017.109. Epub 2018 Jan 15. Nat Rev Genet. 2018. PMID: 29335644 Review.
Cited by
-
Genetic architecture and lifetime dynamics of inbreeding depression in a wild mammal.Nat Commun. 2021 May 20;12(1):2972. doi: 10.1038/s41467-021-23222-9. Nat Commun. 2021. PMID: 34016997 Free PMC article.
-
Nested admixture during and after the Trans-Atlantic Slave Trade on the island of São Tomé.bioRxiv [Preprint]. 2024 Oct 23:2024.10.21.619344. doi: 10.1101/2024.10.21.619344. bioRxiv. 2024. Update in: Mol Biol Evol. 2025 Jul 1;42(7):msaf156. doi: 10.1093/molbev/msaf156. PMID: 39484499 Free PMC article. Updated. Preprint.
-
The influence of gene flow on population viability in an isolated urban caracal population.Mol Ecol. 2024 May;33(9):e17346. doi: 10.1111/mec.17346. Epub 2024 Apr 5. Mol Ecol. 2024. PMID: 38581173 Free PMC article.
-
Genomic consequences of isolation and inbreeding in an island dingo population.bioRxiv [Preprint]. 2024 Mar 12:2023.09.15.557950. doi: 10.1101/2023.09.15.557950. bioRxiv. 2024. Update in: Genome Biol Evol. 2024 Jul 3;16(7):evae130. doi: 10.1093/gbe/evae130. PMID: 37745583 Free PMC article. Updated. Preprint.
-
Parameter Scaling in Population Genetics Simulations may Introduce Unintended Background Selection: Considerations for Scaled Simulation Design.Genome Biol Evol. 2025 May 30;17(6):evaf097. doi: 10.1093/gbe/evaf097. Genome Biol Evol. 2025. PMID: 40405504 Free PMC article.
References
-
- Ceballos F.C., Joshi P.K., Clark D.W., Ramsay M., Wilson J.F. Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet. 2018;19:220–234. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 ES015794/ES/NIEHS NIH HHS/United States
- R01 HL141992/HL/NHLBI NIH HHS/United States
- R01 GM075316/GM/NIGMS NIH HHS/United States
- K01 HL140218/HL/NHLBI NIH HHS/United States
- R01 HL141845/HL/NHLBI NIH HHS/United States
- R01 HG007644/HG/NHGRI NIH HHS/United States
- R01 HL135156/HL/NHLBI NIH HHS/United States
- U01 HG009080/HG/NHGRI NIH HHS/United States
- R01 HL128439/HL/NHLBI NIH HHS/United States
- R21 ES024844/ES/NIEHS NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- P60 MD006902/MD/NIMHD NIH HHS/United States
- RL5 GM118984/GM/NIGMS NIH HHS/United States
- R01 MD010443/MD/NIMHD NIH HHS/United States

