Targeted Repair of p47-CGD in iPSCs by CRISPR/Cas9: Functional Correction without Cleavage in the Highly Homologous Pseudogenes
- PMID: 31543470
- PMCID: PMC6829751
- DOI: 10.1016/j.stemcr.2019.08.008
Targeted Repair of p47-CGD in iPSCs by CRISPR/Cas9: Functional Correction without Cleavage in the Highly Homologous Pseudogenes
Abstract
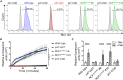
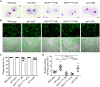
Mutations in the NADPH oxidase, which is crucial for the respiratory burst in phagocytes, result in chronic granulomatous disease (CGD). The only curative treatment option for CGD patients, who suffer from severe infections, is allogeneic bone marrow transplantation. Over 90% of patients with mutations in the p47phox subunit of the oxidase complex carry the deletion c.75_76delGT (ΔGT). This frequent mutation most likely originates via gene conversion from one of the two pseudogenes NCF1B or NCF1C, which are highly homologous to NCF1 (encodes p47phox) but carry the ΔGT mutation. We applied CRISPR/Cas9 to generate patient-like p47-ΔGT iPSCs for disease modeling. To avoid unpredictable chromosomal rearrangements by CRISPR/Cas9-mediated cleavage in the pseudogenes, we developed a gene-correction approach to specifically target NCF1 but leave the pseudogenes intact. Functional assays revealed restored NADPH oxidase activity and killing of bacteria in corrected phagocytes as well as the specificity of this approach.
Keywords: CRISPR/Cas9; NADPH oxidase; NCF1; chronic granulomatous disease (CGD); gene editing; human induced pluripotent stem cells; p47phox; pseudogenes.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Ackermann M., Lachmann N., Hartung S., Eggenschwiler R., Pfaff N., Happle C., Mucci A., Göhring G., Niemann H., Hansen G. Promoter and lineage independent anti-silencing activity of the A2 ubiquitous chromatin opening element for optimized human pluripotent stem cell-based gene therapy. Biomaterials. 2014;35:1531–1542. - PubMed
-
- Güngör T., Teira P., Slatter M., Stussi G., Stepensky P., Moshous D., Vermont C., Ahmad I., Shaw P.J., Telles da Cunha J.M. Reduced-intensity conditioning and HLA-matched haemopoietic stem-cell transplantation in patients with chronic granulomatous disease: a prospective multicentre study. Lancet. 2014;383:436–448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

