Quantification of T-Cell and B-Cell Replication History in Aging, Immunodeficiency, and Newborn Screening
- PMID: 31543882
- PMCID: PMC6730487
- DOI: 10.3389/fimmu.2019.02084
Quantification of T-Cell and B-Cell Replication History in Aging, Immunodeficiency, and Newborn Screening
Abstract
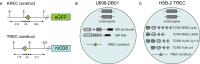
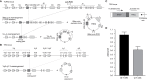
Quantification of T-cell receptor excision circles (TRECs) has impacted on human T-cell research, but interpretations on T-cell replication have been limited due to the lack of a genomic coding joint. We here overcome this limitation with multiplex TRG rearrangement quantification (detecting ~0.98 alleles per TCRαβ+ T cell) and the HSB-2 cell line with a retrovirally introduced TREC construct. We uncovered <5 cell divisions in naive and >10 cell divisions in effector memory T-cell subsets. Furthermore, we show that TREC dilution with age in healthy adults results mainly from increased T cell replication history. This proliferation was significantly increased in patients with predominantly antibody deficiency. Finally, Guthrie cards of neonates with Down syndrome have fewer T and B cells than controls, with similar T-cell and slightly higher B-cell replication. Thus, combined analysis of TRG coding joints and TREC signal joints can be utilized to quantify in vivo T-cell replication, and has direct applications for research into aging, immunodeficiency, and newborn screening.
Keywords: T-cell replication; TREC; TRG; aging; newborn screening; primary immunodeficiency.
Figures




References
-
- Modell V, Orange JS, Quinn J, Modell F. Global report on primary immunodeficiencies: 2018 update from the Jeffrey Modell Centers Network on disease classification, regional trends, treatment modalities, and physician reported outcomes. Immunol Res. (2018) 66:367–80. 10.1007/s12026-018-8996-5 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

