Enhancers Improve the AID-Induced Hypermutation in Episomal Vector for Antibody Affinity Maturation in Mammalian Cell Display
- PMID: 31544892
- PMCID: PMC6698961
- DOI: 10.3390/antib7040042
Enhancers Improve the AID-Induced Hypermutation in Episomal Vector for Antibody Affinity Maturation in Mammalian Cell Display
Abstract
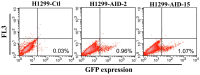
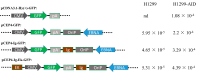
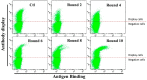
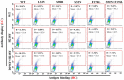
The induction of somatic hypermutation (SHM) in various cell lines by activation-induced cytidine deaminase (AID) has been used in protein-directed selection, especially in antibody affinity maturation. Several antibody affinity maturation systems based on mammalian cells have been developed in recent years, i.e., 293T, H1299, Raji and CHO cells. However, the efficiency of in vitro AID-induced hypermutation is low, restricting the application of such systems. In this study, we examined the role of Ig and Ek enhancers in enhancing SHM in the episomal vector pCEP4 that expresses an anti-high mobility group box 1 (HMGB1) full-length antibody. The plasmid containing the two enhancers exhibited two-fold improvement of mutation rate over pCEP4 in an AID expression H1299 cell line (H1299-AID). With the engineered episomal vector, we improved the affinity of this antibody in H1299-AID cells by 20-fold.
Keywords: activation-induced cytidine deaminase (AID); enhancer; episomal vector; somatic hypermutation (SHM).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Yuan B., Schulz P., Liu R., Sierks M.R. Improved affinity selection using phage display technology and off-rate based selection. Electron. J. Biotechn. 2006;9:171–175. doi: 10.2225/vol9-issue2-fulltext-6. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

