Interbase FRET in RNA: from A to Z
- PMID: 31544922
- PMCID: PMC6821158
- DOI: 10.1093/nar/gkz812
Interbase FRET in RNA: from A to Z
Abstract
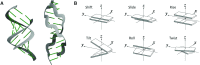
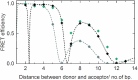
Interbase FRET can reveal highly detailed information about distance, orientation and dynamics in nucleic acids, complementing the existing structure and dynamics techniques. We here report the first RNA base analogue FRET pair, consisting of the donor tCO and the non-emissive acceptor tCnitro. The acceptor ribonucleoside is here synthesised and incorporated into RNA for the first time. This FRET pair accurately reports the average structure of A-form RNA, and its utility for probing RNA structural changes is demonstrated by monitoring the transition from A- to Z-form RNA. Finally, the measured FRET data were compared with theoretical FRET patterns obtained from two previously reported Z-RNA PDB structures, to shed new light on this elusive RNA conformation.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Studying Z-DNA and B- to Z-DNA transitions using a cytosine analogue FRET-pair.Nucleic Acids Res. 2016 Jun 20;44(11):e101. doi: 10.1093/nar/gkw114. Epub 2016 Feb 20. Nucleic Acids Res. 2016. PMID: 26896804 Free PMC article.
-
Characterization of nucleobase analogue FRET acceptor tCnitro.J Phys Chem B. 2010 Jan 21;114(2):1050-6. doi: 10.1021/jp909471b. J Phys Chem B. 2010. PMID: 20039634
-
Toward Complete Sequence Flexibility of Nucleic Acid Base Analogue FRET.J Am Chem Soc. 2017 Jul 12;139(27):9271-9280. doi: 10.1021/jacs.7b04517. Epub 2017 Jun 27. J Am Chem Soc. 2017. PMID: 28613885
-
Multi-fluorophore fluorescence resonance energy transfer for probing nucleic acids structure and folding.Methods Mol Biol. 2006;335:257-71. doi: 10.1385/1-59745-069-3:257. Methods Mol Biol. 2006. PMID: 16785633 Review.
-
Fluorescent nucleobase analogues for base-base FRET in nucleic acids: synthesis, photophysics and applications.Beilstein J Org Chem. 2018 Jan 10;14:114-129. doi: 10.3762/bjoc.14.7. eCollection 2018. Beilstein J Org Chem. 2018. PMID: 29441135 Free PMC article. Review.
Cited by
-
Severe Prolonged Drought Favours Stress-Tolerant Microbes in Australian Drylands.Microb Ecol. 2023 Nov;86(4):3097-3110. doi: 10.1007/s00248-023-02303-w. Epub 2023 Oct 25. Microb Ecol. 2023. PMID: 37878053 Free PMC article.
-
Recognition of non-CpG repeats in Alu and ribosomal RNAs by the Z-RNA binding domain of ADAR1 induces A-Z junctions.Nat Commun. 2021 Feb 4;12(1):793. doi: 10.1038/s41467-021-21039-0. Nat Commun. 2021. PMID: 33542240 Free PMC article.
-
Interbase-FRET binding assay for pre-microRNAs.Sci Rep. 2021 Apr 30;11(1):9396. doi: 10.1038/s41598-021-88922-0. Sci Rep. 2021. PMID: 33931703 Free PMC article.
-
Supramolecular Fluorescence Resonance Energy Transfer in Nucleobase-Modified Fluorogenic RNA Aptamers.Angew Chem Int Ed Engl. 2020 Apr 20;59(17):6760-6764. doi: 10.1002/anie.201916707. Epub 2020 Mar 6. Angew Chem Int Ed Engl. 2020. PMID: 32052536 Free PMC article.
-
Stealth Fluorescence Labeling for Live Microscopy Imaging of mRNA Delivery.J Am Chem Soc. 2021 Apr 14;143(14):5413-5424. doi: 10.1021/jacs.1c00014. Epub 2021 Apr 2. J Am Chem Soc. 2021. PMID: 33797236 Free PMC article.
References
-
- Harpur A.G., Wouters F.S., Bastiaens P.I.H.. Imaging FRET between spectrally similar GFP molecules in single cells. Nat. Biotechnol. 2001; 19:167–169. - PubMed
-
- Hillger F., Hanni D., Nettels D., Geister S., Grandin M., Textor M., Schuler B.. Probing protein-chaperone interactions with single-molecule fluorescence spectroscopy. Angew. Chem. Int. Ed. 2008; 47:6184–6188. - PubMed
-
- Gubaev A., Klostermeier D.. Klostermeier D., Hammann C.. RNA Structure and Folding: Biophysical Techniques and Prediction Methods. 2013; Berlin: De Gruyter; 181–214.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

