Plant Organelle Genome Replication
- PMID: 31546578
- PMCID: PMC6843274
- DOI: 10.3390/plants8100358
Plant Organelle Genome Replication
Abstract
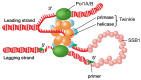
Mitochondria and chloroplasts perform essential functions in respiration, ATP production, and photosynthesis, and both organelles contain genomes that encode only some of the proteins that are required for these functions. The proteins and mechanisms for organelle DNA replication are very similar to bacterial or phage systems. The minimal replisome may consist of DNA polymerase, a primase/helicase, and a single-stranded DNA binding protein (SSB), similar to that found in bacteriophage T7. In Arabidopsis, there are two genes for organellar DNA polymerases and multiple potential genes for SSB, but there is only one known primase/helicase protein to date. Genome copy number varies widely between type and age of plant tissues. Replication mechanisms are only poorly understood at present, and may involve multiple processes, including recombination-dependent replication (RDR) in plant mitochondria and perhaps also in chloroplasts. There are still important questions remaining as to how the genomes are maintained in new organelles, and how genome copy number is determined. This review summarizes our current understanding of these processes.
Keywords: DNA repair; DNA replication; chloroplast DNA; plant mitochondrial DNA; recombination-dependent replication (RDR).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Hooke R. Micrographia: Or Some Physiological Descriptions of Minute Bodies Made by Magnifying Glasses with Observations and Inquiries Thereupon. Science Heritage, Ltd.; Lincolnwood, IL, USA: 1987. 323p Facsimile Edition.
-
- Harris H. The Birth of the Cell. Yale University Press; New Haven, CT, USA: 1999. p. 212.
-
- Schimper A.F.W. Ueber die Entwickelung der Chlorophyllkörner und Farbkörper. Botanische Zeitung. 1883;41:105–160.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

