Heterologous Expression of GbTCP4, a Class II TCP Transcription Factor, Regulates Trichome Formation and Root Hair Development in Arabidopsis
- PMID: 31546783
- PMCID: PMC6771151
- DOI: 10.3390/genes10090726
Heterologous Expression of GbTCP4, a Class II TCP Transcription Factor, Regulates Trichome Formation and Root Hair Development in Arabidopsis
Abstract
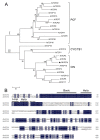
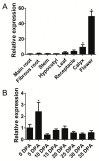
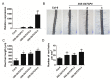
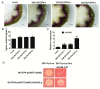
Two class I family teosinte branched1/cycloidea/proliferating cell factor1 (TCP) proteins from allotetraploid cotton are involved in cotton fiber cell differentiation and elongation and root hair development. However, the biological function of most class II TCP proteins is unclear. This study sought to reveal the characteristics and functions of the sea-island cotton class II TCP gene GbTCP4 by biochemical, genetic, and molecular biology methods. GbTCP4 protein localizes to nuclei, binding two types of TCP-binding cis-acting elements, including the one in its promoter. Expression pattern analysis revealed that GbTCP4 is widely expressed in tissues, with the highest level in flowers. GbTCP4 is expressed at different fiber development stages and has high transcription in fibers beginning at 5 days post anthesis (DPA). GbTCP4 overexpression increases primary root hair length and density and leaf and stem trichomes in transgenic Arabidopsis relative to wild-type plants (WT). GbTCP4 binds directly to the CAPRICE (CPC) promoter, increasing CPC transcript levels in roots and reducing them in leaves. Compared with WT plants, lignin content in the stems of transgenic Arabidopsis overexpressing GbTCP4 increased, and AtCAD5 gene transcript levels increased. These results suggest that GbTCP4 regulates trichome formation and root hair development in Arabidopsis and may be a candidate gene for regulating cotton fiber elongation.
Keywords: TCP transcription factor; root hair development; sea-island cotton; trichome formation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Ni Z., Li B., Neumann M.P., LÜ M., Fan L. Isolation and expression analysis of two genes encoding cinnamate 4-hydroxylase from Cotton (Gossypium hirsutum) J. Integr. Agric. 2014;13:2102–2112. doi: 10.1016/S2095-3119(13)60643-7. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

