The Anti-Amyloidogenic Action of Doxycycline: A Molecular Dynamics Study on the Interaction with Aβ42
- PMID: 31546787
- PMCID: PMC6769662
- DOI: 10.3390/ijms20184641
The Anti-Amyloidogenic Action of Doxycycline: A Molecular Dynamics Study on the Interaction with Aβ42
Abstract
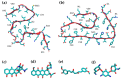
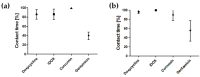
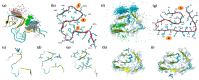
The pathological aggregation of amyloidogenic proteins is a hallmark of many neurological diseases, including Alzheimer's disease and prion diseases. We have shown both in vitro and in vivo that doxycycline can inhibit the aggregation of Aβ42 amyloid fibrils and disassemble mature amyloid fibrils. However, the molecular mechanisms of the drug's anti-amyloidogenic property are not understood. In this study, a series of molecular dynamics simulations were performed to explain the molecular mechanism of the destabilization of Aβ42 fibrils by doxycycline and to compare the action of doxycycline with those of iododoxorubicin (a toxic structural homolog of tetracyclines), curcumin (known to have anti-amyloidogenic activity) and gentamicin (an antibiotic with no experimental evidence of anti-amyloidogenic properties). We found that doxycycline tightly binds the exposed hydrophobic amino acids of the Aβ42 amyloid fibrils, partly leading to destabilization of the fibrillar structure. Clarifying the molecular determinants of doxycycline binding to Aβ42 may help devise further strategies for structure-based drug design for Alzheimer's disease.
Keywords: Alzheimer’s disease; amyloid-beta protein; curcumin; doxycycline; iododoxorubicin; molecular dynamics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Scrutiny of the mechanism of small molecule inhibitor preventing conformational transition of amyloid-β42 monomer: insights from molecular dynamics simulations.J Biomol Struct Dyn. 2018 Feb;36(3):663-678. doi: 10.1080/07391102.2017.1291363. Epub 2017 Feb 28. J Biomol Struct Dyn. 2018. PMID: 28162045
-
Insights into the inhibitory mechanism of a resveratrol and clioquinol hybrid against Aβ42 aggregation and protofibril destabilization: A molecular dynamics simulation study.J Biomol Struct Dyn. 2019 Aug;37(12):3183-3197. doi: 10.1080/07391102.2018.1511475. Epub 2018 Dec 24. J Biomol Struct Dyn. 2019. PMID: 30582723
-
N-Terminus Binding Preference for Either Tanshinone or Analogue in Both Inhibition of Amyloid Aggregation and Disaggregation of Preformed Amyloid Fibrils-Toward Introducing a Kind of Novel Anti-Alzheimer Compounds.ACS Chem Neurosci. 2017 Jul 19;8(7):1577-1588. doi: 10.1021/acschemneuro.7b00080. Epub 2017 Apr 28. ACS Chem Neurosci. 2017. PMID: 28406293
-
Protection mechanisms against Abeta42 aggregation.Curr Alzheimer Res. 2008 Dec;5(6):548-54. doi: 10.2174/156720508786898460. Curr Alzheimer Res. 2008. PMID: 19075581 Review.
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
Cited by
-
Amyloid Disassembly: What Can We Learn from Chaperones?Biomedicines. 2022 Dec 17;10(12):3276. doi: 10.3390/biomedicines10123276. Biomedicines. 2022. PMID: 36552032 Free PMC article. Review.
-
The Use of Antimicrobial and Antiviral Drugs in Alzheimer's Disease.Int J Mol Sci. 2020 Jul 12;21(14):4920. doi: 10.3390/ijms21144920. Int J Mol Sci. 2020. PMID: 32664669 Free PMC article. Review.
-
N-Alkylamino Stilbene Compounds as Amyloid β Inhibitors for Alzheimer's Disease Research.Molecules. 2025 Jun 5;30(11):2471. doi: 10.3390/molecules30112471. Molecules. 2025. PMID: 40509358 Free PMC article.
-
In Silico Analysis of Nanoplastics' and β-amyloid Fibrils' Interactions.Molecules. 2023 Jan 2;28(1):388. doi: 10.3390/molecules28010388. Molecules. 2023. PMID: 36615582 Free PMC article.
-
Small-Molecule Inhibitors of Amyloid Beta: Insights from Molecular Dynamics-Part A: Endogenous Compounds and Repurposed Drugs.Pharmaceuticals (Basel). 2025 Feb 23;18(3):306. doi: 10.3390/ph18030306. Pharmaceuticals (Basel). 2025. PMID: 40143085 Free PMC article. Review.
References
-
- Gianni B.L., Bellotti V., Gianni A.M., Merlini G. New Drug Therapy of Amyloidoses: Resorption of AL-Type Deposits with 4′-Iodo-4′-Deoxydoxorubicin. Blood. 1995;86:855–861. - PubMed
-
- Merlini G., Ascari E., Amboldi N., Bellotti V., Arbustini E., Perfetti V., Ferrari M., Zorzoli I., Marinone M.G., Garini P. Interaction of the anthracycline 4′-iodo-4′-deoxydoxorubicin with amyloid fibrils: Inhibition of amyloidogenesis. Proc. Natl. Acad. Sci. USA. 1995;92:2959–2963. doi: 10.1073/pnas.92.7.2959. - DOI - PMC - PubMed
-
- Tagliavini F., McArthur R.A., Canciani B., Giaccone G., Porro M., Bugiani M., Lievens P.M.J., Bugiani O., Peri E., Dall’Ara P., et al. Effectiveness of anthracycline against experimental prion disease in Syrian hamsters. Science. 1997;276:1119–1122. doi: 10.1126/science.276.5315.1119. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

