MORF9 Functions in Plastid RNA Editing with Tissue Specificity
- PMID: 31546885
- PMCID: PMC6769653
- DOI: 10.3390/ijms20184635
MORF9 Functions in Plastid RNA Editing with Tissue Specificity
Abstract
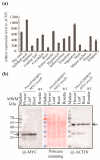
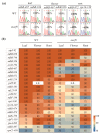
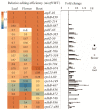
RNA editing in plant mitochondria and plastids converts specific nucleotides from cytidine (C) to uridine (U). These editing events differ among plant species and are relevant to developmental stages or are impacted by environmental conditions. Proteins of the MORF family are essential components of plant editosomes. One of the members, MORF9, is considered the core protein of the editing complex and is involved in the editing of most sites in chloroplasts. In this study, the phenotypes of a T-DNA insertion line with loss of MORF9 and of the genetic complementation line of Arabidopsis were analyzed, and the editing efficiencies of plastid RNAs in roots, rosette leaves, and flowers from the morf9 mutant and the wild-type (WT) control were compared by bulk-cDNA sequencing. The results showed that most of the known MORF9-associated plastid RNA editing events in rosette leaves and flowers were similarly reduced by morf9 mutation, with the exception that the editing rate of the sites ndhB-872 and psbF-65 declined in the leaves and that of ndhB-586 decreased only in the flowers. In the roots, however, the loss of MORF9 had a much lower effect on overall plastid RNA editing, with nine sites showing no significant editing efficiency change, including accD-794, ndhD-383, psbZ-50, ndhF-290, ndhD-878, matK-706, clpP1-559, rpoA-200, and ndhD-674, which were reduced in the other tissues. Furthermore, we found that during plant aging, MORF9 mRNA level, but not the protein level, was downregulated in senescent leaves. On the basis of these observations, we suggest that MORF9-mediated RNA editing is tissue-dependent and the resultant organelle proteomes are pertinent to the specific tissue functions.
Keywords: Arabidopsis thaliana; MORF9; RNA editing; organelles; tissue-specific.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bentolila S., Heller W.P., Sun T., Babina A.M., Friso G., van Wijk K.J., Hanson M.R. Rip1, a member of an arabidopsis protein family, interacts with the protein rare1 and broadly affects rna editing. Proc. Natl. Acad. Sci. USA. 2012;109:E1453–E1461. doi: 10.1073/pnas.1121465109. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

