Forming Big Datasets through Latent Class Concatenation of Imperfectly Matched Databases Features
- PMID: 31546899
- PMCID: PMC6771148
- DOI: 10.3390/genes10090727
Forming Big Datasets through Latent Class Concatenation of Imperfectly Matched Databases Features
Abstract
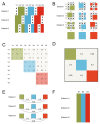
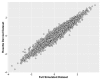
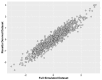
Informatics researchers often need to combine data from many different sources to increase statistical power and study subtle or complicated effects. Perfect overlap of measurements across academic studies is rare since virtually every dataset is collected for a unique purpose and without coordination across parties not-at-hand (i.e., informatics researchers in the future). Thus, incomplete concordance of measurements across datasets poses a major challenge for researchers seeking to combine public databases. In any given field, some measurements are fairly standard, but every organization collecting data makes unique decisions on instruments, protocols, and methods of processing the data. This typically denies literal concatenation of the raw data since constituent cohorts do not have the same measurements (i.e., columns of data). When measurements across datasets are similar prima facie, there is a desire to combine the data to increase power, but mixing non-identical measurements could greatly reduce the sensitivity of the downstream analysis. Here, we discuss a statistical method that is applicable when certain patterns of missing data are found; namely, it is possible to combine datasets that measure the same underlying constructs (or latent traits) when there is only partial overlap of measurements across the constituent datasets. Our method, ROSETTA empirically derives a set of common latent trait metrics for each related measurement domain using a novel variation of factor analysis to ensure equivalence across the constituent datasets. The advantage of combining datasets this way is the simplicity, statistical power, and modeling flexibility of a single joint analysis of all the data. Three simulation studies show the performance of ROSETTA on datasets with only partially overlapping measurements (i.e., systematically missing information), benchmarked to a condition of perfectly overlapped data (i.e., full information). The first study examined a range of correlations, while the second study was modeled after the observed correlations in a well-characterized clinical, behavioral cohort. Both studies consistently show significant correlations >0.94, often >0.96, indicating the robustness of the method and validating the general approach. The third study varied within and between domain correlations and compared ROSETTA to multiple imputation and meta-analysis as two commonly used methods that ostensibly solve the same data integration problem. We provide one alternative to meta-analysis and multiple imputation by developing a method that statistically equates similar but distinct manifest metrics into a set of empirically derived metrics that can be used for analysis across all datasets.
Keywords: data blending; data integration; data pools; databases; informatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Schmitt T.A. Current methodological considerations in exploratory and confirmatory factor analysis. J. Psychoeduc. Assess. 2011;29:304–321. doi: 10.1177/0734282911406653. - DOI
-
- Higham N.J. Computing the nearest correlation matrix—A problem from finance. IMA J. Numer. Anal. 2002;22:329–343. doi: 10.1093/imanum/22.3.329. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

