Hepatic Transcriptomics Reveals that Lipogenesis Is a Key Signaling Pathway in Isocitrate Dehydrogenase 2 Deficient Mice
- PMID: 31546946
- PMCID: PMC6770969
- DOI: 10.3390/genes10090728
Hepatic Transcriptomics Reveals that Lipogenesis Is a Key Signaling Pathway in Isocitrate Dehydrogenase 2 Deficient Mice
Abstract
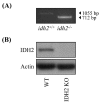
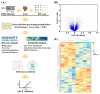
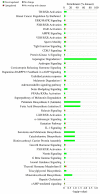
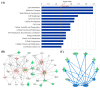
Mitochondrial nicotinamide adenine dinucleotide phosphate (NADP+)-dependent isocitrate dehydrogenase (IDH2) plays a key role in the intermediary metabolism and energy production via catalysing oxidative decarboxylation of isocitrate to α-ketoglutarate in the tricarboxylic acid (TCA) cycle. Despite studies reporting potential interlinks between IDH2 and various diseases, there is lack of effort to comprehensively characterize signature(s) of IDH2 knockout (IDH2 KO) mice. A total of 6583 transcripts were identified from both wild-type (WT) and IDH2 KO mice liver tissues. Afterwards, 167 differentially expressed genes in the IDH2 KO group were short-listed compared to the WT group based on our criteria. The online bioinformatic analyses indicated that lipid metabolism is the most significantly influenced metabolic process in IDH2 KO mice. Moreover, the TR/RXR activation pathway was predicted as the top canonical pathway significantly affected by IDH2 KO. The key transcripts found in the bioinformatic analyses were validated by qPCR analysis, corresponding to the transcriptomics results. Further, an additional qPCR analysis confirmed that IDH2 KO caused a decrease in hepatic de novo lipogenesis via the activation of the fatty acid β-oxidation process. Our unbiased transcriptomics approach and validation experiments suggested that IDH2 might play a key role in homeostasis of lipid metabolism.
Keywords: Idh2; bioinformatics; hepatic transcriptomics; knockout mouse; lipid metabolism.
Conflict of interest statement
The authors declare no competing of interest.
Figures


References
-
- Jo S.H., Son M.K., Koh H.J., Lee S.M., Song I.H., Kim Y.O., Lee Y.S., Jeong K.S., Kim W.B., Park J.W., et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. - DOI - PubMed
-
- Koshland D.E., Jr., Walsh K., LaPorte D.C. Sensitivity of metabolic fluxes to covalent control. Curr. Top. Cell. Regul. 1985;27:13–22. - PubMed
-
- Berg J., Tymoczko J., Stryer L. Biochemistry. 5th ed. W. H. Freeman and Company; New York, NY, USA: 2002. Section 22.4 fatty acids are synthesized and degraded by different pathways; p. 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

