Purine-Metabolising Enzymes and Apoptosis in Cancer
- PMID: 31547393
- PMCID: PMC6769685
- DOI: 10.3390/cancers11091354
Purine-Metabolising Enzymes and Apoptosis in Cancer
Abstract
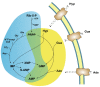
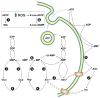
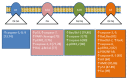
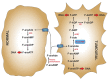
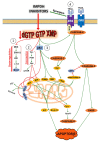
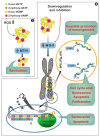
The enzymes of both de novo and salvage pathways for purine nucleotide synthesis are regulated to meet the demand of nucleic acid precursors during proliferation. Among them, the salvage pathway enzymes seem to play the key role in replenishing the purine pool in dividing and tumour cells that require a greater amount of nucleotides. An imbalance in the purine pools is fundamental not only for preventing cell proliferation, but also, in many cases, to promote apoptosis. It is known that tumour cells harbour several mutations that might lead to defective apoptosis-inducing pathways, and this is probably at the basis of the initial expansion of the population of neoplastic cells. Therefore, knowledge of the molecular mechanisms that lead to apoptosis of tumoural cells is key to predicting the possible success of a drug treatment and planning more effective and focused therapies. In this review, we describe how the modulation of enzymes involved in purine metabolism in tumour cells may affect the apoptotic programme. The enzymes discussed are: ectosolic and cytosolic 5'-nucleotidases, purine nucleoside phosphorylase, adenosine deaminase, hypoxanthine-guanine phosphoribosyltransferase, and inosine-5'-monophosphate dehydrogenase, as well as recently described enzymes particularly expressed in tumour cells, such as deoxynucleoside triphosphate triphosphohydrolase and 7,8-dihydro-8-oxoguanine triphosphatase.
Keywords: ADA; CD73; HPRT; IMPDH; MTH1; PNP; SAMHD1; apoptosis; cN-II; purine salvage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

