Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium distachyon
- PMID: 31547557
- PMCID: PMC6843561
- DOI: 10.3390/plants8100362
Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium distachyon
Abstract
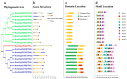
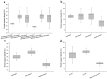
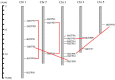
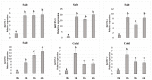
Trehalose biosynthesis enzyme homologues in plants contain two families, trehalose-6-phosphate synthases (TPSs) and trehalose-6-phosphate phosphatases (TPPs). Both families participate in trehalose synthesis and a variety of stress-resistance processes. Here, nine BdTPS and ten BdTPP genes were identified based on the Brachypodium distachyon genome, and all genes were classified into three classes. The Class I and Class II members differed substantially in gene structures, conserved motifs, and protein sequence identities, implying varied gene functions. Gene duplication analysis showed that one BdTPS gene pair and four BdTPP gene pairs are formed by duplication events. The value of Ka/Ks (non-synonymous/synonymous) was less than 1, suggesting purifying selection in these gene families. The cis-elements and gene interaction network prediction showed that many family members may be involved in stress responses. The quantitative real-time reverse transcription (qRT-PCR) results further supported that most BdTPSs responded to at least one stress or abscisic acid (ABA) treatment, whereas over half of BdTPPs were downregulated after stress treatment, implying that BdTPSs play a more important role in stress responses than BdTPPs. This work provides a foundation for the genome-wide identification of the B. distachyon TPS-TPP gene families and a frame for further studies of these gene families in abiotic stress responses.
Keywords: Brachypodium distachyon; TPP; TPS; abiotic stress; expression analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources

