K+ Channel-SEC11 Binding Exchange Regulates SNARE Assembly for Secretory Traffic
- PMID: 31548266
- PMCID: PMC6836825
- DOI: 10.1104/pp.19.00919
K+ Channel-SEC11 Binding Exchange Regulates SNARE Assembly for Secretory Traffic
Abstract
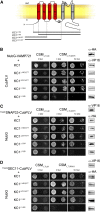
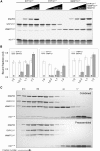
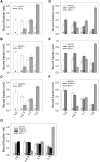
Cell expansion requires that ion transport and secretory membrane traffic operate in concert. Evidence from Arabidopsis (Arabidopsis thaliana) indicates that such coordination is mediated by physical interactions between subsets of so-called SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins, which drive the final stages of vesicle fusion, and K+ channels, which facilitate uptake of the cation to maintain cell turgor pressure as the cell expands. However, the sequence of SNARE binding with the K+ channels and its interweaving within the events of SNARE complex assembly for exocytosis remains unclear. We have combined protein-protein interaction and electrophysiological analyses to resolve the binding interactions of the hetero-oligomeric associations. We find that the RYxxWE motif, located within the voltage sensor of the K+ channels, is a nexus for multiple SNARE interactions. Of these, K+ channel binding and its displacement of the regulatory protein SEC11 is critical to prime the Qa-SNARE SYP121. Our results indicate a stabilizing role for the Qbc-SNARE SNAP33 in the Qa-SNARE transition to SNARE complex assembly with the R-SNARE VAMP721. They also suggest that, on its own, the R-SNARE enters an anomalous binding mode with the channels, possibly as a fail-safe measure to ensure a correct binding sequence. Thus, we suggest that SYP121 binding to the K+ channels serves the role of a primary trigger to initiate assembly of the secretory machinery for exocytosis.
© 2019 The author(s).
Figures







References
-
- Archbold JK, Whitten AE, Hu SH, Collins BM, Martin JL (2014) SNARE-ing the structures of Sec1/Munc18 proteins. Curr Opin Struct Biol 29: 44–51 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/D001528/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C500595/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P12750/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M001601/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N006909/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

