Regulation of Cell Division in Bacteria by Monitoring Genome Integrity and DNA Replication Status
- PMID: 31548275
- PMCID: PMC6941525
- DOI: 10.1128/JB.00408-19
Regulation of Cell Division in Bacteria by Monitoring Genome Integrity and DNA Replication Status
Abstract
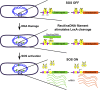
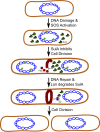
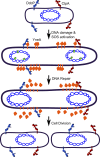
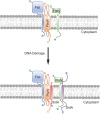
All organisms regulate cell cycle progression by coordinating cell division with DNA replication status. In eukaryotes, DNA damage or problems with replication fork progression induce the DNA damage response (DDR), causing cyclin-dependent kinases to remain active, preventing further cell cycle progression until replication and repair are complete. In bacteria, cell division is coordinated with chromosome segregation, preventing cell division ring formation over the nucleoid in a process termed nucleoid occlusion. In addition to nucleoid occlusion, bacteria induce the SOS response after replication forks encounter DNA damage or impediments that slow or block their progression. During SOS induction, Escherichia coli expresses a cytoplasmic protein, SulA, that inhibits cell division by directly binding FtsZ. After the SOS response is turned off, SulA is degraded by Lon protease, allowing for cell division to resume. Recently, it has become clear that SulA is restricted to bacteria closely related to E. coli and that most bacteria enforce the DNA damage checkpoint by expressing a small integral membrane protein. Resumption of cell division is then mediated by membrane-bound proteases that cleave the cell division inhibitor. Further, many bacterial cells have mechanisms to inhibit cell division that are regulated independently from the canonical LexA-mediated SOS response. In this review, we discuss several pathways used by bacteria to prevent cell division from occurring when genome instability is detected or before the chromosome has been fully replicated and segregated.
Keywords: DNA damage; SOS response; cell cycle; cell division; checkpoint.
Copyright © 2020 American Society for Microbiology.
Figures
Similar articles
-
A replication-inhibited unsegregated nucleoid at mid-cell blocks Z-ring formation and cell division independently of SOS and the SlmA nucleoid occlusion protein in Escherichia coli.J Bacteriol. 2014 Jan;196(1):36-49. doi: 10.1128/JB.01230-12. Epub 2013 Oct 18. J Bacteriol. 2014. PMID: 24142249 Free PMC article.
-
sfi-independent filamentation in Escherichia coli Is lexA dependent and requires DNA damage for induction.J Bacteriol. 1997 Mar;179(6):1931-9. doi: 10.1128/jb.179.6.1931-1939.1997. J Bacteriol. 1997. PMID: 9068638 Free PMC article.
-
Inactivation of Cell Division Protein FtsZ by SulA Makes Lon Indispensable for the Viability of a ppGpp0 Strain of Escherichia coli.J Bacteriol. 2015 Dec 7;198(4):688-700. doi: 10.1128/JB.00693-15. J Bacteriol. 2015. PMID: 26644431 Free PMC article.
-
SosA in Staphylococci: an addition to the paradigm of membrane-localized, SOS-induced cell division inhibition in bacteria.Curr Genet. 2020 Jun;66(3):495-499. doi: 10.1007/s00294-019-01052-z. Epub 2020 Jan 10. Curr Genet. 2020. PMID: 31925496 Review.
-
The divisome at 25: the road ahead.Curr Opin Microbiol. 2017 Apr;36:85-94. doi: 10.1016/j.mib.2017.01.007. Epub 2017 Mar 6. Curr Opin Microbiol. 2017. PMID: 28254403 Free PMC article. Review.
Cited by
-
Redesign of ultrasensitive and robust RecA gene circuit to sense DNA damage.Microb Biotechnol. 2021 Nov;14(6):2481-2496. doi: 10.1111/1751-7915.13767. Epub 2021 Mar 4. Microb Biotechnol. 2021. PMID: 33661573 Free PMC article.
-
Delving Into the Functional Meaning of Phenotypic Variation in Mycobacterial Persistence: Who Benefits the Most From Programmed Death of Individual Cells?Microbiol Insights. 2020 Jul 27;13:1178636120945304. doi: 10.1177/1178636120945304. eCollection 2020. Microbiol Insights. 2020. PMID: 32782432 Free PMC article.
-
CdrS Is a Global Transcriptional Regulator Influencing Cell Division in Haloferax volcanii.mBio. 2021 Aug 31;12(4):e0141621. doi: 10.1128/mBio.01416-21. Epub 2021 Jul 13. mBio. 2021. PMID: 34253062 Free PMC article.
-
Transcriptomic Reprograming of Xanthomonas campestris pv. campestris after Treatment with Hydrolytic Products Derived from Glucosinolates.Plants (Basel). 2021 Aug 11;10(8):1656. doi: 10.3390/plants10081656. Plants (Basel). 2021. PMID: 34451701 Free PMC article.
-
Queuosine biosynthetic enzyme, QueE moonlights as a cell division regulator.PLoS Genet. 2024 May 20;20(5):e1011287. doi: 10.1371/journal.pgen.1011287. eCollection 2024 May. PLoS Genet. 2024. PMID: 38768229 Free PMC article.
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC, p 463–497.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

