Experimental evolution of immunological specificity
- PMID: 31548373
- PMCID: PMC6789748
- DOI: 10.1073/pnas.1904828116
Experimental evolution of immunological specificity
Abstract
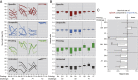
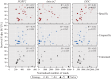
Memory and specificity are hallmarks of the adaptive immune system. Contrary to prior belief, innate immune systems can also provide forms of immune memory, such as immune priming in invertebrates and trained immunity in vertebrates. Immune priming can even be specific but differs remarkably in cellular and molecular functionality from the well-studied adaptive immune system of vertebrates. To date, it is unknown whether and how the level of specificity in immune priming can adapt during evolution in response to natural selection. We tested the evolution of priming specificity in an invertebrate model, the beetle Tribolium castaneum Using controlled evolution experiments, we selected beetles for either specific or unspecific immune priming toward the bacteria Pseudomonas fluorescens, Lactococcus lactis, and 4 strains of the entomopathogen Bacillus thuringiensis After 14 generations of host selection, specificity of priming was not universally higher in the lines selected for specificity, but rather depended on the bacterium used for priming and challenge. The insect pathogen B. thuringiensis induced the strongest priming effect. Differences between the evolved populations were mirrored in the transcriptomic response, revealing involvement of immune, metabolic, and transcription-modifying genes. Finally, we demonstrate that the induction strength of a set of differentially expressed immune genes predicts the survival probability of the evolved lines upon infection. We conclude that high specificity of immune priming can evolve rapidly for certain bacteria, most likely due to changes in the regulation of immune genes.
Keywords: immune memory; immune priming; immunological specificity; innate immunity; trained immunity.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Murphy K. P., Janeway C. A., Mowat A., Janeway’s Immunobiology (Garland Science, New York, ed. 8, 2012).
-
- Netea M. G., Quintin J., van der Meer J. W. M., Trained immunity: A memory for innate host defense. Cell Host Microbe 9, 355–361 (2011). - PubMed
-
- Reimer-Michalski E.-M., Conrath U., Innate immune memory in plants. Semin. Immunol. 28, 319–327 (2016). - PubMed
-
- Milutinović B., Kurtz J., Immune memory in invertebrates. Semin. Immunol. 28, 328–342 (2016). - PubMed
-
- Kurtz J., Specific memory within innate immune systems. Trends Immunol. 26, 186–192 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

