Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1
- PMID: 31548395
- PMCID: PMC6789920
- DOI: 10.1073/pnas.1906663116
Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1
Abstract
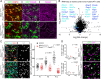
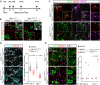
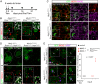
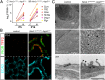
The extraordinarily thin alveolar type 1 (AT1) cell constitutes nearly the entire gas exchange surface and allows passive diffusion of oxygen into the blood stream. Despite such an essential role, the transcriptional network controlling AT1 cells remains unclear. Using cell-specific knockout mouse models, genomic profiling, and 3D imaging, we found that NK homeobox 2-1 (Nkx2-1) is expressed in AT1 cells and is required for the development and maintenance of AT1 cells. Without Nkx2-1, developing AT1 cells lose 3 defining features-molecular markers, expansive morphology, and cellular quiescence-leading to alveolar simplification and lethality. NKX2-1 is also cell-autonomously required for the same 3 defining features in mature AT1 cells. Intriguingly, Nkx2-1 mutant AT1 cells activate gastrointestinal (GI) genes and form dense microvilli-like structures apically. Single-cell RNA-seq supports a linear transformation of Nkx2-1 mutant AT1 cells toward a GI fate. Whole lung ChIP-seq shows NKX2-1 binding to 68% of genes that are down-regulated upon Nkx2-1 deletion, including 93% of known AT1 genes, but near-background binding to up-regulated genes. Our results place NKX2-1 at the top of the AT1 cell transcriptional hierarchy and demonstrate remarkable plasticity of an otherwise terminally differentiated cell type.
Keywords: NK homeobox 2-1; alveolar type 1 cell; alveologenesis; lung development; transcriptional control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo.Nat Commun. 2021 May 4;12(1):2509. doi: 10.1038/s41467-021-22817-6. Nat Commun. 2021. PMID: 33947861 Free PMC article.
-
PRDM3/16 regulate chromatin accessibility required for NKX2-1 mediated alveolar epithelial differentiation and function.Nat Commun. 2024 Sep 16;15(1):8112. doi: 10.1038/s41467-024-52154-3. Nat Commun. 2024. PMID: 39284798 Free PMC article.
-
The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation.J Biol Chem. 2012 Oct 26;287(44):37282-95. doi: 10.1074/jbc.M112.408401. Epub 2012 Sep 6. J Biol Chem. 2012. PMID: 22955271 Free PMC article.
-
Homeobox genes in cardiovascular development.Curr Top Dev Biol. 1998;40:1-44. doi: 10.1016/s0070-2153(08)60363-4. Curr Top Dev Biol. 1998. PMID: 9673847 Review.
-
Thyroid Transcription Factor-1: Structure, Expression, Function and Its Relationship with Disease.Biomed Res Int. 2021 Sep 28;2021:9957209. doi: 10.1155/2021/9957209. eCollection 2021. Biomed Res Int. 2021. PMID: 34631891 Free PMC article. Review.
Cited by
-
Development of human alveolar epithelial cell models to study distal lung biology and disease.iScience. 2022 Jan 15;25(2):103780. doi: 10.1016/j.isci.2022.103780. eCollection 2022 Feb 18. iScience. 2022. PMID: 35169685 Free PMC article.
-
Phosphorylated nuclear DICER1 promotes open chromatin state and lineage plasticity of AT2 tumor cells in lung adenocarcinomas.Sci Adv. 2023 Jul 28;9(30):eadf6210. doi: 10.1126/sciadv.adf6210. Epub 2023 Jul 26. Sci Adv. 2023. PMID: 37494452 Free PMC article.
-
p53 governs an AT1 differentiation programme in lung cancer suppression.Nature. 2023 Jul;619(7971):851-859. doi: 10.1038/s41586-023-06253-8. Epub 2023 Jul 19. Nature. 2023. PMID: 37468633 Free PMC article.
-
Epithelial Yap/Taz are required for functional alveolar regeneration following acute lung injury.JCI Insight. 2023 Sep 7;8(19):e173374. doi: 10.1172/jci.insight.173374. JCI Insight. 2023. PMID: 37676731 Free PMC article.
-
CEBPA restricts alveolar type 2 cell plasticity during development and injury-repair.Nat Commun. 2024 May 16;15(1):4148. doi: 10.1038/s41467-024-48632-3. Nat Commun. 2024. PMID: 38755149 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

