RNA ligation precedes the retrotransposition of U6/LINE-1 chimeric RNA
- PMID: 31548405
- PMCID: PMC6789731
- DOI: 10.1073/pnas.1805404116
RNA ligation precedes the retrotransposition of U6/LINE-1 chimeric RNA
Abstract
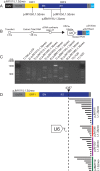
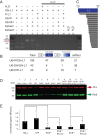
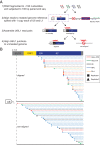
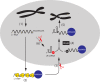
Long interspersed element-1 (LINE-1 or L1) amplifies via retrotransposition. Active L1s encode 2 proteins (ORF1p and ORF2p) that bind their encoding transcript to promote retrotransposition in cis The L1-encoded proteins also promote the retrotransposition of small-interspersed element RNAs, noncoding RNAs, and messenger RNAs in trans Some L1-mediated retrotransposition events consist of a copy of U6 RNA conjoined to a variably 5'-truncated L1, but how U6/L1 chimeras are formed requires elucidation. Here, we report the following: The RNA ligase RtcB can join U6 RNAs ending in a 2',3'-cyclic phosphate to L1 RNAs containing a 5'-OH in vitro; depletion of endogenous RtcB in HeLa cell extracts reduces U6/L1 RNA ligation efficiency; retrotransposition of U6/L1 RNAs leads to U6/L1 pseudogene formation; and a unique cohort of U6/L1 chimeric RNAs are present in multiple human cell lines. Thus, these data suggest that U6 small nuclear RNA (snRNA) and RtcB participate in the formation of chimeric RNAs and that retrotransposition of chimeric RNA contributes to interindividual genetic variation.
Keywords: LINE-1; RNA ligation; RtcB; U6 snRNA; retrotransposon.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: J.V.M. is an inventor on patent US6150160, is a paid consultant for Gilead Sciences and a privately held company founded by Flagship Pioneering, and is on the American Society of Human Genetics Board of Directors.
Figures

References
-
- Lander E. S., et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Erratum in: Nature411, 720 (2001) and Nature412, 565 (2001). - PubMed
-
- Smit A. F., The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6, 743–748 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

