Epitope Sequences in Dengue Virus NS1 Protein Identified by Monoclonal Antibodies
- PMID: 31548529
- PMCID: PMC6698852
- DOI: 10.3390/antib6040014
Epitope Sequences in Dengue Virus NS1 Protein Identified by Monoclonal Antibodies
Abstract
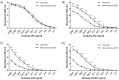
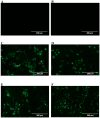
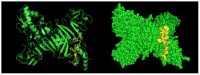
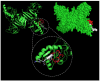
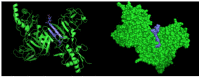
Dengue nonstructural protein 1 (NS1) is a multi-functional glycoprotein with essential functions both in viral replication and modulation of host innate immune responses. NS1 has been established as a good surrogate marker for infection. In the present study, we generated four anti-NS1 monoclonal antibodies against recombinant NS1 protein from dengue virus serotype 2 (DENV2), which were used to map three NS1 epitopes. The sequence 193AVHADMGYWIESALNDT209 was recognized by monoclonal antibodies 2H5 and 4H1BC, which also cross-reacted with Zika virus (ZIKV) protein. On the other hand, the sequence 25VHTWTEQYKFQPES38 was recognized by mAb 4F6 that did not cross react with ZIKV. Lastly, a previously unidentified DENV2 NS1-specific epitope, represented by the sequence 127ELHNQTFLIDGPETAEC143, is described in the present study after reaction with mAb 4H2, which also did not cross react with ZIKV. The selection and characterization of the epitope, specificity of anti-NS1 mAbs, may contribute to the development of diagnostic tools able to differentiate DENV and ZIKV infections.
Keywords: NS1; Zika virus; amino acid sequences; antibody recognition; dengue virus; mAbs.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures
Similar articles
-
Development and characterization of mouse monoclonal antibodies against monomeric dengue virus non-structural glycoprotein 1 (NS1).J Virol Methods. 2015 Sep 15;222:214-23. doi: 10.1016/j.jviromet.2015.06.003. Epub 2015 Jun 9. J Virol Methods. 2015. PMID: 26070890
-
ZIKV-Specific NS1 Epitopes as Serological Markers of Acute Zika Virus Infection.J Infect Dis. 2019 Jun 19;220(2):203-212. doi: 10.1093/infdis/jiz092. J Infect Dis. 2019. PMID: 30901054
-
Cross-reactive antibodies targeting surface-exposed non-structural protein 1 (NS1) of dengue virus-infected cells recognize epitopes on the spaghetti loop of the β-ladder domain.PLoS One. 2022 May 26;17(5):e0266136. doi: 10.1371/journal.pone.0266136. eCollection 2022. PLoS One. 2022. PMID: 35617160 Free PMC article.
-
Development and characterization of serotype-specific monoclonal antibodies against the dengue virus-4 (DENV-4) non-structural protein (NS1).Virol J. 2018 Feb 6;15(1):30. doi: 10.1186/s12985-018-0925-7. Virol J. 2018. PMID: 29409531 Free PMC article.
-
Delayed and highly specific antibody response to nonstructural protein 1 (NS1) revealed during natural human ZIKV infection by NS1-based capture ELISA.BMC Infect Dis. 2018 Jun 14;18(1):275. doi: 10.1186/s12879-018-3173-y. BMC Infect Dis. 2018. PMID: 29898684 Free PMC article.
Cited by
-
Adaptive Immunity to Dengue Virus: Slippery Slope or Solid Ground for Rational Vaccine Design?Pathogens. 2020 Jun 15;9(6):470. doi: 10.3390/pathogens9060470. Pathogens. 2020. PMID: 32549226 Free PMC article. Review.
-
Flavivirus Cross-Reactivity to Dengue Nonstructural Protein 1 Antigen Detection Assays.Diagnostics (Basel). 2019 Dec 24;10(1):11. doi: 10.3390/diagnostics10010011. Diagnostics (Basel). 2019. PMID: 31878299 Free PMC article.
-
Special Issue: Monoclonal Antibodies.Antibodies (Basel). 2018 Apr 10;7(2):17. doi: 10.3390/antib7020017. Antibodies (Basel). 2018. PMID: 31544869 Free PMC article.
-
Flavivirus NS1 and Its Potential in Vaccine Development.Vaccines (Basel). 2021 Jun 9;9(6):622. doi: 10.3390/vaccines9060622. Vaccines (Basel). 2021. PMID: 34207516 Free PMC article. Review.
-
Anti-DENV-NS1 monoclonal antibody for the differential histopathological diagnosis of hemorrhagic fever caused by dengue.Braz J Microbiol. 2022 Jun;53(2):777-783. doi: 10.1007/s42770-022-00697-2. Epub 2022 Feb 7. Braz J Microbiol. 2022. PMID: 35129818 Free PMC article.
References
-
- Secretaria de Vigilância em Saúde, Ministério da Saúde. Bol. Epidemiol. 2017;48:1–9.
Grants and funding
- 2009/53894-3/Fundação de Amparo à Pesquisa do Estado de São Paulo
- 2011/51761-6/Fundação de Amparo à Pesquisa do Estado de São Paulo
- 2014/17595-0/Fundação de Amparo à Pesquisa do Estado de São Paulo
- 2013/06589-6/Fundação de Amparo à Pesquisa do Estado de São Paulo
- 2013/50955-7/Fundação de Amparo à Pesquisa do Estado de São Paulo
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

