Gene Ontology Causal Activity Modeling (GO-CAM) moves beyond GO annotations to structured descriptions of biological functions and systems
- PMID: 31548717
- PMCID: PMC7012280
- DOI: 10.1038/s41588-019-0500-1
Gene Ontology Causal Activity Modeling (GO-CAM) moves beyond GO annotations to structured descriptions of biological functions and systems
Abstract
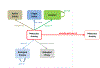
To increase the utility of Gene Ontology annotations for interpretation of genome-wide experimental data, we have developed GO-CAM, a structured framework for linking multiple GO annotations into an integrated model of a biological system. We expect that GO-CAM will enable new applications in pathway and network analysis as well as improving standard GO annotations for traditional GO-based applications.
Figures


References
-
- Mungall CJ, Dietze H, Osumi-Sutherland D. Use of OWL within the gene ontology. BioRxiv 2014, October:010090.
-
- Anindya R, Aygün O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase nedd4, but not cockayne syndrome proteins or BRCA1. Mol Cell 2007, November 9;28(3):386–97. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

