The long noncoding RNA NEAT1_1 is seemingly dispensable for normal tissue homeostasis and cancer cell growth
- PMID: 31551298
- PMCID: PMC6859857
- DOI: 10.1261/rna.071456.119
The long noncoding RNA NEAT1_1 is seemingly dispensable for normal tissue homeostasis and cancer cell growth
Abstract
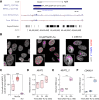
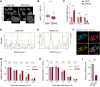
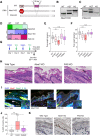
NEAT1 is one of the most studied lncRNAs, in part because its silencing in mice causes defects in mammary gland development and corpus luteum formation and protects them from skin cancer development. Moreover, depleting NEAT1 in established cancer cell lines reduces growth and sensitizes cells to DNA damaging agents. However, NEAT1 produces two isoforms and because the short isoform, NEAT1_1, completely overlaps the 5' part of the long NEAT1_2 isoform; the respective contributions of each of the isoforms to these phenotypes has remained unclear. Whereas NEAT1_1 is highly expressed in most tissues, NEAT1_2 is the central architectural component of paraspeckles, which are nuclear bodies that assemble in specific tissues and cells exposed to various forms of stress. Using dual RNA-FISH to detect both NEAT1_1 outside of the paraspeckles and NEAT1_2/NEAT1 inside this nuclear body, we report herein that NEAT1_1 levels are dynamically regulated during the cell cycle and targeted for degradation by the nuclear RNA exosome. Unexpectedly, however, cancer cells engineered to lack NEAT1_1, but not NEAT1_2, do not exhibit cell cycle defects. Moreover, Neat1_1-specific knockout mice do not exhibit the phenotypes observed in Neat1-deficient mice. We propose that NEAT1 functions are mainly, if not exclusively, attributable to NEAT1_2 and, by extension, to paraspeckles.
Keywords: NEAT1 isoforms; RNA exosome; cell cycle; lncRNA; mouse genetics; paraspeckles.
© 2019 Adriaens et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, et al. 2016. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med 22: 861–868. 10.1038/nm.4135 - DOI - PubMed
-
- Ahmed ASI, Dong K, Liu J, Wen T, Yu L, Xu F, Kang X, Osman I, Hu G, Bunting KM, et al. 2018. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc Natl Acad Sci 115: E8660–E8667. 10.1073/pnas.1803725115 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
