RNA detection with high specificity and sensitivity using nested fluorogenic Mango NASBA
- PMID: 31551299
- PMCID: PMC6859864
- DOI: 10.1261/rna.072629.119
RNA detection with high specificity and sensitivity using nested fluorogenic Mango NASBA
Abstract
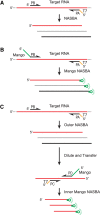
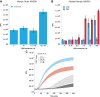
There is a pressing need for nucleic acid-based assays that are capable of rapidly and reliably detecting pathogenic organisms. Many of the techniques available for the detection of pathogenic RNA possess one or more limiting factors that make the detection of low-copy RNA challenging. Although RT-PCR is the most commonly used method for detecting pathogen-related RNA, it requires expensive thermocycling equipment and is comparatively slow. Isothermal methods promise procedural simplicity but have traditionally suffered from amplification artifacts that tend to preclude easy identification of target nucleic acids. Recently, the isothermal SHERLOCK system overcame this problem by using CRISPR to distinguish amplified target sequences from artifactual background signal. However, this system comes at the cost of introducing considerable enzymatic complexity and a corresponding increase in total assay time. Therefore, simpler and less expensive strategies are highly desirable. Here, we demonstrate that by nesting NASBA primers and modifying the NASBA inner primers to encode an RNA Mango aptamer sequence we can dramatically increase the sensitivity of NASBA to 1.5 RNA molecules per microliter. As this isothermal nucleic acid detection scheme directly produces a fluorescent reporter, real-time detection is intrinsic to the assay. Nested Mango NASBA is highly specific and, in contrast to existing RNA detection systems, offers a cheap, simple, and specific way to rapidly detect single-molecule amounts of pathogenic RNA.
Keywords: Mango; RNA; TO1 fluorophore; isothermal amplification; nested NASBA; nucleic acid detection; single-molecule detection.
© 2019 Abdolahzadeh et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
References
-
- Alam KK, Jung JK, Verosloff MS, Clauer PR, Lee JW, Capdevila DA, Pastén PA, Giedroc DP, Collins JJ, Lucks JB. 2019. Rapid, low-cost detection of water contaminants using regulated in vitro transcription. Synth Biol. 10.1101/619296 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
