Resistance Evolution against Phage Combinations Depends on the Timing and Order of Exposure
- PMID: 31551330
- PMCID: PMC6759759
- DOI: 10.1128/mBio.01652-19
Resistance Evolution against Phage Combinations Depends on the Timing and Order of Exposure
Abstract
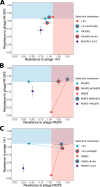
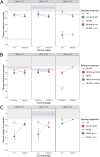
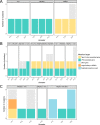
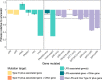
Phage therapy is a promising alternative to chemotherapeutic antibiotics for the treatment of bacterial infections. However, despite recent clinical uses of combinations of phages to treat multidrug-resistant infections, a mechanistic understanding of how bacteria evolve resistance against multiple phages is lacking, limiting our ability to deploy phage combinations optimally. Here, we show, using Pseudomonas aeruginosa and pairs of phages targeting shared or distinct surface receptors, that the timing and order of phage exposure determine the strength, cost, and mutational basis of resistance. Whereas sequential exposure allowed bacteria to acquire multiple resistance mutations effective against both phages, this evolutionary trajectory was prevented by simultaneous exposure, resulting in quantitatively weaker resistance. The order of phage exposure determined the fitness costs of sequential resistance, such that certain sequential orders imposed much higher fitness costs than the same phage pair in the reverse order. Together, these data suggest that phage combinations can be optimized to limit the strength of evolved resistances while maximizing their associated fitness costs to promote the long-term efficacy of phage therapy.IMPORTANCE Globally rising rates of antibiotic resistance have renewed interest in phage therapy where combinations of phages have been successfully used to treat multidrug-resistant infections. To optimize phage therapy, we first need to understand how bacteria evolve resistance against combinations of multiple phages. Here, we use simple laboratory experiments and genome sequencing to show that the timing and order of phage exposure determine the strength, cost, and mutational basis of resistance evolution in the opportunistic pathogen Pseudomonas aeruginosa These findings suggest that phage combinations can be optimized to limit the emergence and persistence of resistance, thereby promoting the long-term usefulness of phage therapy.
Keywords: Pseudomonas aeruginosa; bacteriophage therapy; bacteriophages; evolutionary biology; resistance evolution.
Copyright © 2019 Wright et al.
Figures
Similar articles
-
Optimizing bacteriophage treatment of resistant Pseudomonas.mSphere. 2024 Jul 30;9(7):e0070723. doi: 10.1128/msphere.00707-23. Epub 2024 Jun 27. mSphere. 2024. PMID: 38934592 Free PMC article.
-
Multi-strain phage induced clearance of bacterial infections.PLoS Comput Biol. 2025 Feb 4;21(2):e1012793. doi: 10.1371/journal.pcbi.1012793. eCollection 2025 Feb. PLoS Comput Biol. 2025. PMID: 39903766 Free PMC article.
-
Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae.Appl Environ Microbiol. 2012 Aug;78(16):5646-52. doi: 10.1128/AEM.00757-12. Epub 2012 Jun 1. Appl Environ Microbiol. 2012. PMID: 22660719 Free PMC article.
-
Bacteriophages of Pseudomonas aeruginosa: long-term prospects for use in phage therapy.Adv Virus Res. 2014;88:227-78. doi: 10.1016/B978-0-12-800098-4.00005-2. Adv Virus Res. 2014. PMID: 24373314 Review.
-
Phage against the Machine: The SIE-ence of Superinfection Exclusion.Viruses. 2024 Aug 23;16(9):1348. doi: 10.3390/v16091348. Viruses. 2024. PMID: 39339825 Free PMC article. Review.
Cited by
-
The genetic basis of phage susceptibility, cross-resistance and host-range in Salmonella.Microbiology (Reading). 2021 Dec;167(12):001126. doi: 10.1099/mic.0.001126. Microbiology (Reading). 2021. PMID: 34910616 Free PMC article.
-
Phage therapy for Klebsiella pneumoniae: Understanding bacteria-phage interactions for therapeutic innovations.PLoS Pathog. 2025 Apr 8;21(4):e1012971. doi: 10.1371/journal.ppat.1012971. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40198880 Free PMC article. Review.
-
The Basis for Natural Multiresistance to Phage in Pseudomonas aeruginosa.Antibiotics (Basel). 2020 Jun 18;9(6):339. doi: 10.3390/antibiotics9060339. Antibiotics (Basel). 2020. PMID: 32570896 Free PMC article.
-
Semantics Count in the Description of the Interactions Between Bacteria and Bacteriophage.Phage (New Rochelle). 2025 Mar 17;6(1):3-4. doi: 10.1089/phage.2024.0063. eCollection 2025 Mar. Phage (New Rochelle). 2025. PMID: 40291344 No abstract available.
-
Optimizing bacteriophage treatment of resistant Pseudomonas.mSphere. 2024 Jul 30;9(7):e0070723. doi: 10.1128/msphere.00707-23. Epub 2024 Jun 27. mSphere. 2024. PMID: 38934592 Free PMC article.
References
-
- Review on Antimicrobial Resistance. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance, United Kingdom.
-
- Rohde C, Resch G, Pirnay J-P, Blasdel B, Debarbieux L, Gelman D, Górski A, Hazan R, Huys I, Kakabadze E, Łobocka M, Maestri A, Almeida GMDF, Makalatia K, Malik D, Mašlaňová I, Merabishvili M, Pantucek R, Rose T, Štveráková D, Van Raemdonck H, Verbeken G, Chanishvili N. 2018. Expert opinion on three phage therapy related topics: bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 10:E178. doi:10.3390/v10040178. - DOI - PMC - PubMed
-
- Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59:145–155. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

