Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments
- PMID: 31551353
- PMCID: PMC6827292
- DOI: 10.1074/jbc.REV119.009178
Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments
Abstract
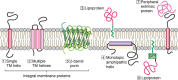
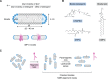
A third of the genes in prokaryotic and eukaryotic genomes encode membrane proteins that are either essential for signal transduction and solute transport or function as scaffold structures. Unlike many of their soluble counterparts, the overall structural and functional organization of membrane proteins is sparingly understood. Recent advances in X-ray crystallography, cryo-EM, and nuclear magnetic resonance (NMR) are closing this gap by enabling an in-depth view of these ever-elusive proteins at atomic resolution. Despite substantial technological advancements, however, the overall proportion of membrane protein entries in the Protein Data Bank (PDB) remains <4%. This paucity is mainly attributed to difficulties associated with their expression and purification, propensity to form large multisubunit complexes, and challenges pertinent to identification of an ideal detergent, lipid, or detergent/lipid mixture that closely mimic their native environment. NMR is a powerful technique to obtain atomic-resolution and dynamic details of a protein in solution. This is accomplished through an assortment of isotopic labeling schemes designed to acquire multiple spectra that facilitate deduction of the final protein structure. In this review, we discuss current approaches and technological developments in the determination of membrane protein structures by solution NMR and highlight recent structural and mechanistic insights gained with this technique. We also discuss strategies for overcoming size limitations in NMR applications, and we explore a plethora of membrane mimetics available for the structural and mechanistic understanding of these essential cellular proteins.
Keywords: G-protein–coupled receptor (GPCR); amphipol; bicelle; membrane mimetic; membrane protein; micelle; nanodisc; nanotechnology; nuclear magnetic resonance (NMR); structural biology.
© 2019 Puthenveetil and Vinogradova.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Ernst R. R., Bodenhausen G., and Wokaun A. (1987) Principles of Nuclear Magnetic Resonance in One and Two Dimensions, Oxford University Press, Oxford, UK
-
- Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, New York
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

