Mice depleted for Exchange Proteins Directly Activated by cAMP (Epac) exhibit irregular liver regeneration in response to partial hepatectomy
- PMID: 31551444
- PMCID: PMC6760117
- DOI: 10.1038/s41598-019-50219-8
Mice depleted for Exchange Proteins Directly Activated by cAMP (Epac) exhibit irregular liver regeneration in response to partial hepatectomy
Abstract
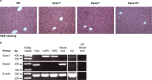
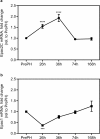
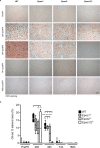
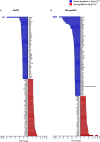
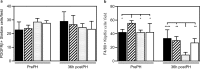
The exchange proteins directly activated by cAMP 1 and 2 (Epac1 and Epac2) are expressed in a cell specific manner in the liver, but their biological functions in this tissue are poorly understood. The current study was undertaken to begin to determine the potential roles of Epac1 and Epac2 in liver physiology and disease. Male C57BL/6J mice in which expression of Epac1 and/or Epac2 are deleted, were subjected to partial hepatectomy and the regenerating liver was analyzed with regard to lipid accumulation, cell replication and protein expression. In response to partial hepatectomy, deletion of Epac1 and/or Epac2 led to increased hepatocyte proliferation 36 h post surgery, and the transient steatosis observed in wild type mice was virtually absent in mice lacking both Epac1 and Epac2. The expression of the protein cytochrome P4504a14, which is implicated in hepatic steatosis and fibrosis, was substantially reduced upon deletion of Epac1/2, while a number of factors involved in lipid metabolism were significantly decreased. Moreover, the number of Küpffer cells was affected, and Epac2 expression was increased in the liver of wild type mice in response to partial hepatectomy, further supporting a role for these proteins in liver function. This study establishes hepatic phenotypic abnormalities in mice deleted for Epac1/2 for the first time, and introduces Epac1/2 as regulators of hepatocyte proliferation and lipid accumulation in the regenerative process.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Berthet J, Rall TW, Sutherland EW. The relationship of epinephrine and glucagon to liver phosphorylase. IV. Effect of epinephrine and glucagon on the reactivation of phosphorylase in liver homogenates. The Journal of biological chemistry. 1957;224:463–475. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

