Fungal Planet description sheets: 868-950
- PMID: 31551622
- PMCID: PMC6712538
- DOI: 10.3767/persoonia.2019.42.11
Fungal Planet description sheets: 868-950
Abstract
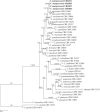
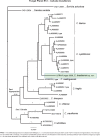
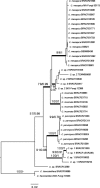
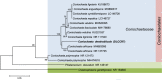
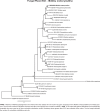
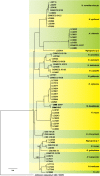
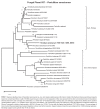
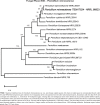
Novel species of fungi described in this study include those from various countries as follows: Australia, Chaetomella pseudocircinoseta and Coniella pseudodiospyri on Eucalyptus microcorys leaves, Cladophialophora eucalypti, Teratosphaeria dunnii and Vermiculariopsiella dunnii on Eucalyptus dunnii leaves, Cylindrium grande and Hypsotheca eucalyptorum on Eucalyptus grandis leaves, Elsinoe salignae on Eucalyptus saligna leaves, Marasmius lebeliae on litter of regenerating subtropical rainforest, Phialoseptomonium eucalypti (incl. Phialoseptomonium gen. nov.) on Eucalyptus grandis × camaldulensis leaves, Phlogicylindrium pawpawense on Eucalyptus tereticornis leaves, Phyllosticta longicauda as an endophyte from healthy Eustrephus latifolius leaves, Pseudosydowia eucalyptorum on Eucalyptus sp. leaves, Saitozyma wallum on Banksia aemula leaves, Teratosphaeria henryi on Corymbia henryi leaves. Brazil, Aspergillus bezerrae, Backusella azygospora, Mariannaea terricola and Talaromyces pernambucoensis from soil, Calonectria matogrossensis on Eucalyptus urophylla leaves, Calvatia brasiliensis on soil, Carcinomyces nordestinensis on Bromelia antiacantha leaves, Dendryphiella stromaticola on small branches of an unidentified plant, Nigrospora brasiliensis on Nopalea cochenillifera leaves, Penicillium alagoense as a leaf endophyte on a Miconia sp., Podosordaria nigrobrunnea on dung, Spegazzinia bromeliacearum as a leaf endophyte on Tilandsia catimbauensis, Xylobolus brasiliensis on decaying wood. Bulgaria, Kazachstania molopis from the gut of the beetle Molops piceus. Croatia, Mollisia endocrystallina from a fallen decorticated Picea abies tree trunk. Ecuador, Hygrocybe rodomaculata on soil. Hungary, Alfoldia vorosii (incl. Alfoldia gen. nov.) from Juniperus communis roots, Kiskunsagia ubrizsyi (incl. Kiskunsagia gen. nov.) from Fumana procumbens roots. India, Aureobasidium tremulum as laboratory contaminant, Leucosporidium himalayensis and Naganishia indica from windblown dust on glaciers. Italy, Neodevriesia cycadicola on Cycas sp. leaves, Pseudocercospora pseudomyrticola on Myrtus communis leaves, Ramularia pistaciae on Pistacia lentiscus leaves, Neognomoniopsis quercina (incl. Neognomoniopsis gen. nov.) on Quercus ilex leaves. Japan, Diaporthe fructicola on Passiflora edulis × P. edulis f. flavicarpa fruit, Entoloma nipponicum on leaf litter in a mixed Cryptomeria japonica and Acer spp. forest. Macedonia, Astraeus macedonicus on soil. Malaysia, Fusicladium eucalyptigenum on Eucalyptus sp. twigs, Neoacrodontiella eucalypti (incl. Neoacrodontiella gen. nov.) on Eucalyptus urophylla leaves. Mozambique, Meliola gorongosensis on dead Philenoptera violacea leaflets. Nepal, Coniochaeta dendrobiicola from Dendriobium lognicornu roots. New Zealand, Neodevriesia sexualis and Thozetella neonivea on Archontophoenix cunninghamiana leaves. Norway, Calophoma sandfjordenica from a piece of board on a rocky shoreline, Clavaria parvispora on soil, Didymella finnmarkica from a piece of Pinus sylvestris driftwood. Poland, Sugiyamaella trypani from soil. Portugal, Colletotrichum feijoicola from Acca sellowiana. Russia, Crepidotus tobolensis on Populus tremula debris, Entoloma ekaterinae, Entoloma erhardii and Suillus gastroflavus on soil, Nakazawaea ambrosiae from the galleries of Ips typographus under the bark of Picea abies. Slovenia, Pluteus ludwigii on twigs of broadleaved trees. South Africa, Anungitiomyces stellenboschiensis (incl. Anungitiomyces gen. nov.) and Niesslia stellenboschiana on Eucalyptus sp. leaves, Beltraniella pseudoportoricensis on Podocarpus falcatus leaf litter, Corynespora encephalarti on Encephalartos sp. leaves, Cytospora pavettae on Pavetta revoluta leaves, Helminthosporium erythrinicola on Erythrina humeana leaves, Helminthosporium syzygii on a Syzygium sp. bark canker, Libertasomyces aloeticus on Aloe sp. leaves, Penicillium lunae from Musa sp. fruit, Phyllosticta lauridiae on Lauridia tetragona leaves, Pseudotruncatella bolusanthi (incl. Pseudotruncatellaceae fam. nov.) and Dactylella bolusanthi on Bolusanthus speciosus leaves. Spain, Apenidiella foetida on submerged plant debris, Inocybe grammatoides on Quercus ilex subsp. ilex forest humus, Ossicaulis salomii on soil, Phialemonium guarroi from soil. Thailand, Pantospora chromolaenae on Chromolaena odorata leaves. Ukraine, Cadophora helianthi from Helianthus annuus stems. USA, Boletus pseudopinophilus on soil under slash pine, Botryotrichum foricae, Penicillium americanum and Penicillium minnesotense from air. Vietnam, Lycoperdon vietnamense on soil. Morphological and culture characteristics are supported by DNA barcodes.
Keywords: ITS nrDNA barcodes; LSU; new taxa; systematics.
Figures





































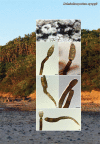



























































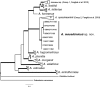



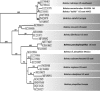

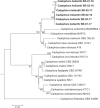




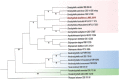




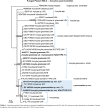




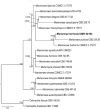



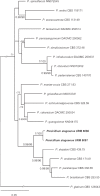



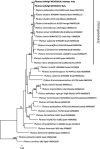
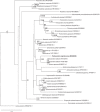




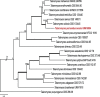
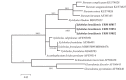
References
-
- Abdalla MY, Al-Rokibah AA. 2003. New record of the pyrenomycete Coniochaeta velutina causing leaf blight of date palm. Journal of King Saud University, Agricultural Sciences 15: 177–184.
-
- Aebi B. 1972. Untersuchungen über Discomyceten aus der Gruppe Tapesia-Trichobelonium. Nova Hedwigia 23: 49–112.
-
- Agustí-Brisach C, Gramaje D, García-Jiménez J, et al. 2013. Detection of black-foot and Petri disease pathogens in natural soils of grapevine nurseries and vineyards using bait plants. Plant and Soil 364: 5–13.
-
- Alfenas RF, Pereira OL, Ferreira MA, et al. 2013. Calonectria metrosideri, a highly aggressive pathogen causing leaf blight, root rot, and wilt of Metrosideros spp. in Brazil. Forest Pathology 43: 257–265.
LinkOut - more resources
Full Text Sources
Miscellaneous
