An Outbreak of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Wenzhou, China
- PMID: 31552210
- PMCID: PMC6736603
- DOI: 10.3389/fpubh.2019.00229
An Outbreak of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Wenzhou, China
Abstract
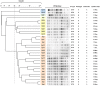
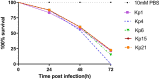
Carbapenem-resistant, hypervirulent Klebsiella pneumoniae (CR-hvKP) has recently emerged as a significant threat to public health. In this study, 29 K. pneumoniae isolates were isolated from eight patients admitted to the intensive care unit (ICU) of a comprehensive teaching hospital located in China from March 2017 to January 2018. Clinical information of patients was the basis for the further analyses of the isolates including antimicrobial susceptibility tests, identification of antibiotic resistance and virulence gene determinants, multilocus sequence typing (MLST), XbaI-macrorestriction by pulsed-field gel electrophoresis (PFGE). Selected isolates representing distinct resistance profiles and virulence phenotypes were screened for hypervirulence in a Galleria mellonella larvae infection model. In the course of the outbreak, the overall mortality rate of patients was 100% (n = 8) attributed to complications arising from CR-hvKP infections. All isolates except one (28/29, 96.6%) were resistant to multiple antimicrobial agents, and harbored diverse resistance determinants that included the globally prevalent carbapenemase bla KPC-2. Most isolates had hypervirulent genotypes being positive for 19 virulence-associated genes, including iutA (25/29, 86.2%), rmpA (27/29, 93.1%), ybtA (27/29, 93.1%), entB (29/29, 100%), fimH (29/29, 100%), and mrkD (29/29, 100%). MLST revealed ST11 for the majority of isolates (26/29, 89,7%). Infection assays demonstrated high mortality in the Galleria mellonella model with the highest LD50 values for three isolates (<105 CFU/mL) demonstrating the degree of hypervirulence of these CR-hvKP isolates, and is discussed relative to previous outbreaks of CR-hvKP.
Keywords: Klebsiella pneumoniae; ST11; carbapenem-resistance; epidemiology; hypervirulent.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

