Moving Mountains-The BRCA1 Promotion of DNA Resection
- PMID: 31552267
- PMCID: PMC6733915
- DOI: 10.3389/fmolb.2019.00079
Moving Mountains-The BRCA1 Promotion of DNA Resection
Abstract
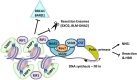
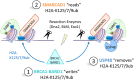
DNA double-strand breaks (DSBs) occur in our cells in the context of chromatin. This type of lesion is toxic, entirely preventing genome continuity and causing cell death or terminal arrest. Several repair mechanisms can act on DNA surrounding a DSB, only some of which carry a low risk of mutation, so that which repair process is utilized is critical to the stability of genetic material of cells. A key component of repair outcome is the degree of DNA resection directed to either side of the break site. This in turn determines the subsequent forms of repair in which DNA homology plays a part. Here we will focus on chromatin and chromatin-bound complexes which constitute the "mountains" that block resection, with a particular focus on how the breast and ovarian cancer predisposition protein-1 (BRCA1) contributes to repair outcomes through overcoming these blocks.
Keywords: 53BP1; BRCA1; SMARCAD1; USP48; homologous recombination; resection.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

