Discovery of Inhibitors for Proliferating Cell Nuclear Antigen Using a Computational-Based Linked-Multiple-Fragment Screen
- PMID: 31552364
- PMCID: PMC6751697
- DOI: 10.1021/acsomega.9b02079
Discovery of Inhibitors for Proliferating Cell Nuclear Antigen Using a Computational-Based Linked-Multiple-Fragment Screen
Abstract
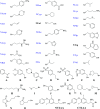
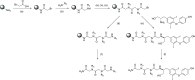
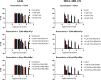
Proliferating cell nuclear antigen (PCNA) is a central factor in DNA replication and repair pathways that plays an essential role in genome stability. The functional roles of PCNA are mediated through an extensive list of protein-protein interactions, each of which transmits specific information in protein assemblies. The flexibility at the PCNA-protein interaction interfaces offers opportunities for the discovery of functionally selective inhibitors of DNA repair pathways. Current fragment-based drug design methodologies can be limited by the flexibility of protein interfaces. These factors motivated an approach to defining compounds that could leverage previously identified subpockets on PCNA that are suitable for fragment-binding sites. Methodologies for screening multiple connected fragment-binding events in distinct subpockets are deployed to improve the selection of fragment combinations. A flexible backbone based on N-alkyl-glycine amides offers a scaffold to combinatorically link multiple fragments for in silico screening libraries that explore the diversity of subpockets at protein interfaces. This approach was applied to discover new potential inhibitors of DNA replication and repair that target PCNA in a multiprotein recognition site. The screens of the libraries were designed to computationally filter ligands based upon the fragments and positions to <1%, which were synthesized and tested for direct binding to PCNA. Molecular dynamics simulations also revealed distinct features of these novel molecules that block key PCNA-protein interactions. Furthermore, a Bayesian classifier predicted 15 of the 16 new inhibitors to be modulators of protein-protein interactions, demonstrating the method's utility as an effective screening filter. The cellular activities of example ligands with similar affinity for PCNA demonstrate unique properties for novel selective synergy with therapeutic DNA-damaging agents in drug-resistant contexts.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
