NPHP proteins are binding partners of nucleoporins at the base of the primary cilium
- PMID: 31553752
- PMCID: PMC6760808
- DOI: 10.1371/journal.pone.0222924
NPHP proteins are binding partners of nucleoporins at the base of the primary cilium
Abstract
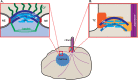
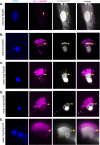
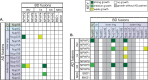
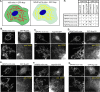
Cilia are microtubule-based organelles that protrude from the surface of eukaryotic cells to generate motility and to sense and respond to environmental cues. In order to carry out these functions, the complement of proteins in the cilium must be specific for the organelle. Regulation of protein entry into primary cilia has been shown to utilize mechanisms and components of nuclear gating, including nucleoporins of the nuclear pore complex (NPC). We show that nucleoporins also localize to the base of motile cilia on the surface of trachea epithelial cells. How nucleoporins are anchored at the cilium base has been unclear as transmembrane nucleoporins, which anchor nucleoporins at the nuclear envelope, have not been found to localize at the cilium. Here we use the directed yeast two-hybrid assay to identify direct interactions between nucleoporins and nephronophthisis proteins (NPHPs) which localize to the cilium base and contribute to cilium assembly and identity. We validate NPHP-nucleoporin interactions in mammalian cells using the knocksideways assay and demonstrate that the interactions occur at the base of the primary cilium using bimolecular fluorescence complementation. We propose that NPHP proteins anchor nucleoporins at the base of primary cilia to regulate protein entry into the organelle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

