Causes and Consequences of A Glutamine Induced Normoxic HIF1 Activity for the Tumor Metabolism
- PMID: 31554283
- PMCID: PMC6802203
- DOI: 10.3390/ijms20194742
Causes and Consequences of A Glutamine Induced Normoxic HIF1 Activity for the Tumor Metabolism
Abstract
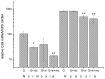
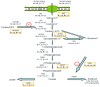
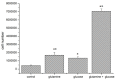
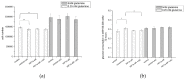
The transcription factor hypoxia-inducible factor 1 (HIF1) is the crucial regulator of genes that are involved in metabolism under hypoxic conditions, but information regarding the transcriptional activity of HIF1 in normoxic metabolism is limited. Different tumor cells were treated under normoxic and hypoxic conditions with various drugs that affect cellular metabolism. HIF1α was silenced by siRNA in normoxic/hypoxic tumor cells, before RNA sequencing and bioinformatics analyses were performed while using the breast cancer cell line MDA-MB-231 as a model. Differentially expressed genes were further analyzed and validated by qPCR, while the activity of the metabolites was determined by enzyme assays. Under normoxic conditions, HIF1 activity was significantly increased by (i) glutamine metabolism, which was associated with the release of ammonium, and it was decreased by (ii) acetylation via acetyl CoA synthetase (ACSS2) or ATP citrate lyase (ACLY), respectively, and (iii) the presence of L-ascorbic acid, citrate, or acetyl-CoA. Interestingly, acetylsalicylic acid, ibuprofen, L-ascorbic acid, and citrate each significantly destabilized HIF1α only under normoxia. The results from the deep sequence analyses indicated that, in HIF1-siRNA silenced MDA-MB-231 cells, 231 genes under normoxia and 1384 genes under hypoxia were transcriptionally significant deregulated in a HIF1-dependent manner. Focusing on glycolysis genes, it was confirmed that HIF1 significantly regulated six normoxic and 16 hypoxic glycolysis-associated gene transcripts. However, the results from the targeted metabolome analyses revealed that HIF1 activity affected neither the consumption of glucose nor the release of ammonium or lactate; however, it significantly inhibited the release of the amino acid alanine. This study comprehensively investigated, for the first time, how normoxic HIF1 is stabilized, and it analyzed the possible function of normoxic HIF1 in the transcriptome and metabolic processes of tumor cells in a breast cancer cell model. Furthermore, these data imply that HIF1 compensates for the metabolic outcomes of glutaminolysis and, subsequently, the Warburg effect might be a direct consequence of the altered amino acid metabolism in tumor cells.
Keywords: HIF1α; L-ascorbic acid; Warburg effect; acetylsalicylic acid; glutamine; glutaminolysis; glycolysis; hypoxia; metabolism; normoxia; tumor.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures










References
-
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

