Genome-Wide Analysis of Left Ventricular Image-Derived Phenotypes Identifies Fourteen Loci Associated With Cardiac Morphogenesis and Heart Failure Development
- PMID: 31554410
- PMCID: PMC6791514
- DOI: 10.1161/CIRCULATIONAHA.119.041161
Genome-Wide Analysis of Left Ventricular Image-Derived Phenotypes Identifies Fourteen Loci Associated With Cardiac Morphogenesis and Heart Failure Development
Abstract
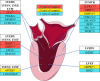
Background: The genetic basis of left ventricular (LV) image-derived phenotypes, which play a vital role in the diagnosis, management, and risk stratification of cardiovascular diseases, is unclear at present.
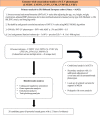
Methods: The LV parameters were measured from the cardiovascular magnetic resonance studies of the UK Biobank. Genotyping was done using Affymetrix arrays, augmented by imputation. We performed genome-wide association studies of 6 LV traits-LV end-diastolic volume, LV end-systolic volume, LV stroke volume, LV ejection fraction, LV mass, and LV mass to end-diastolic volume ratio. The replication analysis was performed in the MESA study (Multi-Ethnic Study of Atherosclerosis). We identified the candidate genes at genome-wide significant loci based on the evidence from extensive bioinformatic analyses. Polygenic risk scores were constructed from the summary statistics of LV genome-wide association studies to predict the heart failure events.
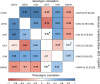
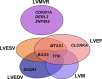
Results: The study comprised 16 923 European UK Biobank participants (mean age 62.5 years; 45.8% men) without prevalent myocardial infarction or heart failure. We discovered 14 genome-wide significant loci (3 loci each for LV end-diastolic volume, LV end-systolic volume, and LV mass to end-diastolic volume ratio; 4 loci for LV ejection fraction, and 1 locus for LV mass) at a stringent P<1×10-8. Three loci were replicated at Bonferroni significance and 7 loci at nominal significance (P<0.05 with concordant direction of effect) in the MESA study (n=4383). Follow-up bioinformatic analyses identified 28 candidate genes that were enriched in the cardiac developmental pathways and regulation of the LV contractile mechanism. Eight genes (TTN, BAG3, GRK5, HSPB7, MTSS1, ALPK3, NMB, and MMP11) supported by at least 2 independent lines of in silico evidence were implicated in the cardiac morphogenesis and heart failure development. The polygenic risk scores of LV phenotypes were predictive of heart failure in a holdout UK Biobank sample of 3106 cases and 224 134 controls (odds ratio 1.41, 95% CI 1.26 - 1.58, for the top quintile versus the bottom quintile of the LV end-systolic volume risk score).
Conclusions: We report 14 genetic loci and indicate several candidate genes that not only enhance our understanding of the genetic architecture of prognostically important LV phenotypes but also shed light on potential novel therapeutic targets for LV remodeling.
Keywords: genome-wide association study; heart failure; left ventricle.
Figures

References
-
- Snipelisky D, Chaudhry SP, Stewart GC. The many faces of heart failure. Card Electrophysiol Clin. 2019;11:11–20. doi: 10.1016/j.ccep.2018.11.001. - PubMed
-
- Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. - PubMed
-
- Grothues F, Smith GC, Moon JC, Bellenger NG, Collins P, Klein HU, Pennell DJ. Comparison of interstudy reproducibility of cardiovascular magnetic resonance with two-dimensional echocardiography in normal subjects and in patients with heart failure or left ventricular hypertrophy. Am J Cardiol. 2002;90:29–34. doi: 10.1016/s0002-9149(02)02381-0. - PubMed
-
- Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. - PubMed
Publication types
MeSH terms
Grants and funding
- N01 HC095163/HC/NHLBI NIH HHS/United States
- HHSN263201500003I/NH/NIH HHS/United States
- N01 HC095168/HC/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- N01 HC095162/HC/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- N01 HC095165/HC/NHLBI NIH HHS/United States
- N02 HL64278/HL/NHLBI NIH HHS/United States
- N01 HC095169/HC/NHLBI NIH HHS/United States
- MR/L016311/1/MRC_/Medical Research Council/United Kingdom
- N01 HC095161/HC/NHLBI NIH HHS/United States
- N01 HC095166/HC/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01 HC095167/HC/NHLBI NIH HHS/United States
- N01 HC095164/HC/NHLBI NIH HHS/United States
- 203553/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- N01 HC095160/HC/NHLBI NIH HHS/United States
- N01 HC095159/HC/NHLBI NIH HHS/United States
- PG/14/89/31194/BHF_/British Heart Foundation/United Kingdom
- P30 DK063491/DK/NIDDK NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

