Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary
- PMID: 31554966
- PMCID: PMC7643931
- DOI: 10.1038/s41586-019-1598-0
Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary
Abstract
Organogenesis is a complex and interconnected process that is orchestrated by multiple boundary tissue interactions1-7. However, it remains unclear how individual, neighbouring components coordinate to establish an integral multi-organ structure. Here we report the continuous patterning and dynamic morphogenesis of hepatic, biliary and pancreatic structures, invaginating from a three-dimensional culture of human pluripotent stem cells. The boundary interactions between anterior and posterior gut spheroids differentiated from human pluripotent stem cells enables retinoic acid-dependent emergence of hepato-biliary-pancreatic organ domains specified at the foregut-midgut boundary organoids in the absence of extrinsic factors. Whereas transplant-derived tissues are dominated by midgut derivatives, long-term-cultured microdissected hepato-biliary-pancreatic organoids develop into segregated multi-organ anlages, which then recapitulate early morphogenetic events including the invagination and branching of three different and interconnected organ structures, reminiscent of tissues derived from mouse explanted foregut-midgut culture. Mis-segregation of multi-organ domains caused by a genetic mutation in HES1 abolishes the biliary specification potential in culture, as seen in vivo8,9. In sum, we demonstrate that the experimental multi-organ integrated model can be established by the juxtapositioning of foregut and midgut tissues, and potentially serves as a tractable, manipulatable and easily accessible model for the study of complex human endoderm organogenesis.
Conflict of interest statement
Figures

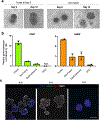
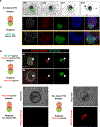
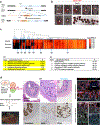
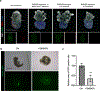


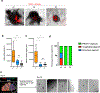





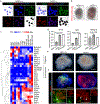
References
-
- Smith DM, Nielsen C, Tabin CJ & Roberts DJ Roles of BMP signaling and Nkx2.5 in patterning at the chick midgut-foregut boundary. Development 127, 3671–3681 (2000). - PubMed
-
- Hentsch B et al. Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev 10, 70–79 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

