Analysis of the B cell receptor repertoire in six immune-mediated diseases
- PMID: 31554970
- PMCID: PMC6795535
- DOI: 10.1038/s41586-019-1595-3
Analysis of the B cell receptor repertoire in six immune-mediated diseases
Abstract
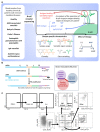
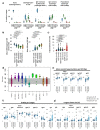
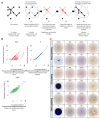
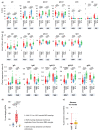
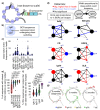
B cells are important in the pathogenesis of many, and perhaps all, immune-mediated diseases. Each B cell expresses a single B cell receptor (BCR)1, and the diverse range of BCRs expressed by the total B cell population of an individual is termed the 'BCR repertoire'. Our understanding of the BCR repertoire in the context of immune-mediated diseases is incomplete, and defining this could provide new insights into pathogenesis and therapy. Here, we compared the BCR repertoire in systemic lupus erythematosus, anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis, Crohn's disease, Behçet's disease, eosinophilic granulomatosis with polyangiitis, and immunoglobulin A (IgA) vasculitis by analysing BCR clonality, use of immunoglobulin heavy-chain variable region (IGHV) genes and-in particular-isotype use. An increase in clonality in systemic lupus erythematosus and Crohn's disease that was dominated by the IgA isotype, together with skewed use of the IGHV genes in these and other diseases, suggested a microbial contribution to pathogenesis. Different immunosuppressive treatments had specific and distinct effects on the repertoire; B cells that persisted after treatment with rituximab were predominately isotype-switched and clonally expanded, whereas the inverse was true for B cells that persisted after treatment with mycophenolate mofetil. Our comparative analysis of the BCR repertoire in immune-mediated disease reveals a complex B cell architecture, providing a platform for understanding pathological mechanisms and designing treatment strategies.
Figures








Comment in
-
Characterizing the BCR repertoire in immune-mediated diseases.Nat Rev Rheumatol. 2020 Jan;16(1):7-8. doi: 10.1038/s41584-019-0339-y. Nat Rev Rheumatol. 2020. PMID: 31780792 Free PMC article.
References
-
- Nossal GJV, Lederberg J. Antibody production by single cells. Nature. 1958;181:1419–1420. - PubMed
-
- Lydyard PM, Whelan A, Fanger MW. Instant Notes Series; Instant Notes in Immunology. 2000;i-x:1–318.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

