Boosting Heterologous Phenazine Production in Pseudomonas putida KT2440 Through the Exploration of the Natural Sequence Space
- PMID: 31555229
- PMCID: PMC6722869
- DOI: 10.3389/fmicb.2019.01990
Boosting Heterologous Phenazine Production in Pseudomonas putida KT2440 Through the Exploration of the Natural Sequence Space
Abstract
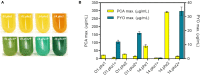
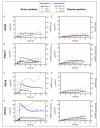
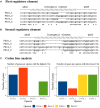
Phenazine-1-carboxylic acid (PCA) and its derivative pyocyanin (PYO) are natural redox mediators in bioelectrochemical systems and have the potential to enable new bioelectrochemical production strategies. The native producer Pseudomonas aeruginosa harbors two identically structured operons in its genome, which encode the enzymes responsible for PCA synthesis [phzA1-G1 (operon 1), phzA2-G2 (operon 2)]. To optimize heterologous phenazines production in the biotech host Pseudomonas putida KT2440, we compared PCA production from both operons originating from P. aeruginosa strain PAO1 (O1.phz1 and O1.phz2) as well as from P. aeruginosa strain PA14 (14.phz1 and 14.phz2). Comparisons of phenazine synthesis and bioelectrochemical activity were performed between heterologous constructs with and without the combination with the genes phzM and phzS required to convert PCA to PYO. Despite a high amino acid homology of all enzymes of more than 97%, P. putida harboring 14.phz2 produced 4-times higher PCA concentrations (80 μg/mL), which resulted in 3-times higher current densities (12 μA/cm2) compared to P. putida 14.phz1. The respective PCA/PYO producer containing the 14.phz2 operon was the best strain with 80 μg/mL PCA, 11 μg/mL PYO, and 22 μA/cm2 current density. Tailoring phenazine production also resulted in improved oxygen-limited metabolic activity of the bacterium through enhanced anodic electron discharge. To elucidate the reason for this superior performance, a detailed structure comparison of the PCA-synthesizing proteins has been performed. The here presented characterization and optimization of these new strains will be useful to improve electroactivity in P. putida for oxygen-limited biocatalysis.
Keywords: PCA; Pseudomonas putida; bioelectrochemical systems; heterologous production; phenazine; pyocyanin.
Figures

References
-
- Bajracharya S., Sharma M., Mohanakrishna G., Dominguez Benneton X., Strik D. P. B. T. B., Sarma P. M., et al. (2016). An overview on emerging bioelectrochemical systems (BESs): technology for sustainable electricity, waste remediation, resource recovery, chemical production and beyond. Renew. Energ. 98 153–170. 10.1016/j.renene.2016.03.002 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

