Gotta Trace 'em All: A Mini-Review on Tools and Procedures for Segmenting Single Neurons Toward Deciphering the Structural Connectome
- PMID: 31555642
- PMCID: PMC6727034
- DOI: 10.3389/fbioe.2019.00202
Gotta Trace 'em All: A Mini-Review on Tools and Procedures for Segmenting Single Neurons Toward Deciphering the Structural Connectome
Abstract
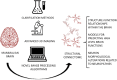
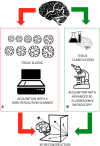
Decoding the morphology and physical connections of all the neurons populating a brain is necessary for predicting and studying the relationships between its form and function, as well as for documenting structural abnormalities in neuropathies. Digitizing a complete and high-fidelity map of the mammalian brain at the micro-scale will allow neuroscientists to understand disease, consciousness, and ultimately what it is that makes us humans. The critical obstacle for reaching this goal is the lack of robust and accurate tools able to deal with 3D datasets representing dense-packed cells in their native arrangement within the brain. This obliges neuroscientist to manually identify the neurons populating an acquired digital image stack, a notably time-consuming procedure prone to human bias. Here we review the automatic and semi-automatic algorithms and software for neuron segmentation available in the literature, as well as the metrics purposely designed for their validation, highlighting their strengths and limitations. In this direction, we also briefly introduce the recent advances in tissue clarification that enable significant improvements in both optical access of neural tissue and image stack quality, and which could enable more efficient segmentation approaches. Finally, we discuss new methods and tools for processing tissues and acquiring images at sub-cellular scales, which will require new robust algorithms for identifying neurons and their sub-structures (e.g., spines, thin neurites). This will lead to a more detailed structural map of the brain, taking twenty-first century cellular neuroscience to the next level, i.e., the Structural Connectome.
Keywords: 3D neuron segmentation; CLARITY; segmentation algorithm; single-cell segmentation; structural connectome.
Figures
References
-
- Ascoli G. A. (2009). The DIADEM Challenge. Available online at: http://diademchallenge.org/ (accessed December 31, 2012).
Publication types
LinkOut - more resources
Full Text Sources

