Nucleosome Remodeling by Fun30SMARCAD1 in the DNA Damage Response
- PMID: 31555662
- PMCID: PMC6737033
- DOI: 10.3389/fmolb.2019.00078
Nucleosome Remodeling by Fun30SMARCAD1 in the DNA Damage Response
Abstract
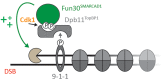
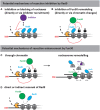
Many cellular pathways are dedicated to maintain the integrity of the genome. In eukaryotes, the underlying DNA transactions occur in the context of chromatin. Cells utilize chromatin and its dynamic nature to regulate those genome integrity pathways. Accordingly, chromatin becomes restructured and modified around DNA damage sites. Here, we review the current knowledge of a chromatin remodeler Fun30SMARCAD1, which plays a key role in genome maintenance. Fun30SMARCAD1 promotes DNA end resection and the repair of DNA double-stranded breaks (DSBs). Notably, however, Fun30SMARCAD1 plays additional roles in maintaining heterochromatin and promoting transcription. Overall, Fun30SMARCAD1 is involved in distinct processes and the specific roles of Fun30SMARCAD1 at DSBs, replication forks and sites of transcription appear discordant at first view. Nonetheless, a picture emerges in which commonalities within these context-dependent roles of Fun30SMARCAD1 exist, which may help to gain a more global understanding of chromatin alterations induced by Fun30SMARCAD1.
Keywords: DNA double-stranded break; DNA end resection; Fun30/SMARCAD1; cell cycle; genome stability; nucleosome remodeling; post-translational modification.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

