Generating the Blood Exposome Database Using a Comprehensive Text Mining and Database Fusion Approach
- PMID: 31557052
- PMCID: PMC6794490
- DOI: 10.1289/EHP4713
Generating the Blood Exposome Database Using a Comprehensive Text Mining and Database Fusion Approach
Abstract
Background: Blood chemicals are routinely measured in clinical or preclinical research studies to diagnose diseases, assess risks in epidemiological research, or use metabolomic phenotyping in response to treatments. A vast volume of blood-related literature is available via the PubMed database for data mining.
Objectives: We aimed to generate a comprehensive blood exposome database of endogenous and exogenous chemicals associated with the mammalian circulating system through text mining and database fusion.
Methods: Using NCBI resources, we retrieved PubMed abstracts, PubChem chemical synonyms, and PMC supplementary tables. We then employed text mining and PubChem crowdsourcing to associate phrases relating to blood with PubChem chemicals. False positives were removed by a phrase pattern and a compound exclusion list.
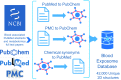
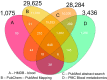
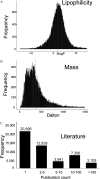
Results: A query to identify blood-related publications in the PubMed database yielded 1.1 million papers. Matching a total of 15 million synonyms from 6.5 million relevant PubChem chemicals against all blood-related publications yielded 37,514 chemicals and 851,999 publications records. Mapping PubChem compound identifiers to the PubMed database yielded 49,940 unique chemicals linked to 676,643 papers. Analysis of open-access metabolomics papers related to blood phrases in the PMC database yielded 4,039 unique compounds and 204 papers. Consolidating these three approaches summed up to a total of 41,474 achiral structures that were linked to 65,957 PubChem CIDs and to over 878,966 PubMed articles. We mapped these compounds to 50 databases such as those covering metabolites and pathways, governmental and toxicological databases, pharmacology resources, and bioassay repositories. In comparison, HMDB, the Human Metabolome Database, links 1,075 compounds to blood-related primary publications.
Conclusion: This new Blood Exposome Database can be used for prioritizing chemicals for systematic reviews, developing target assays in exposome research, identifying compounds in untargeted mass spectrometry, and biological interpretation in metabolomics data. The database is available at http://bloodexposome.org. https://doi.org/10.1289/EHP4713.
Figures
References
-
- Andra SS, Austin C, Patel D, Dolios G, Awawda M, Arora M. 2017. Trends in the application of high-resolution mass spectrometry for human biomonitoring: an analytical primer to studying the environmental chemical space of the human exposome. Environ Int 100:32–61, PMID: 28062070, 10.1016/j.envint.2016.11.026. - DOI - PMC - PubMed
-
- Blaženović I, Kind T, Torbašinović H, Obrenović S, Mehta SS, Tsugawa H, et al. 2017. Comprehensive comparison of in silico MS/MS fragmentation tools of the CASMI contest: Database boosting is needed to achieve 93% accuracy. J Cheminform 9(1):32, PMID: 29086039, 10.1186/s13321-017-0219-x. - DOI - PMC - PubMed
