PML hyposumoylation is responsible for the resistance of pancreatic cancer
- PMID: 31557059
- PMCID: PMC6902661
- DOI: 10.1096/fj.201901091R
PML hyposumoylation is responsible for the resistance of pancreatic cancer
Abstract
The dismal prognosis of pancreatic ductal adenocarcinoma (PDAC) is mainly due to its rapidly acquired resistance to all conventional treatments. Despite drug-specific mechanisms of resistance, none explains how these cells resist the stress induced by any kind of anticancer treatment. Activation of stress-response pathways relies on the post-translational modifications (PTMs) of involved proteins. Among all PTMs, those mediated by the ubiquitin family of proteins play a central role. Our aim was to identify alterations of ubiquitination, neddylation, and sumoylation associated with the multiresistant phenotype and demonstrate their implications in the survival of PDAC cells undergoing treatment. This approach pointed at an alteration of promyelocytic leukemia (PML) protein sumoylation associated with both gemcitabine and oxaliplatin resistance. We could show that this alteration of PML sumoylation is part of a general mechanism of drug resistance, which in addition involves the abnormal activation of NF-κB and cAMP response element binding pathways. Importantly, using patient-derived tumors and cell lines, we identified a correlation between the levels of PML expression and sumoylation and the sensitivity of tumors to anticancer treatments.-Swayden, M., Alzeeb, G., Masoud, R., Berthois, Y., Audebert, S., Camoin, L., Hannouche, L., Vachon, H., Gayet, O., Bigonnet, M., Roques, J., Silvy, F., Carrier, A., Dusetti, N., Iovanna, J. L., Soubeyran, P. PML hyposumoylation is responsible for the resistance of pancreatic cancer.
Keywords: chemoresistance; post-translational modification; promyelocytic leukemia protein; sumoylation; ubiquitin family.
Conflict of interest statement
The authors thank Dr. Arkaitz Carracedo (Center for Cooperative Research in Bioscience, Spain) for providing PML constructs and to Dr. Guillaume Bossis (IGMM, Montpellier, France) for providing anti-SUMO1 and anti-SUMO2/3 hybridomas. High-throughput sequencing was performed at the Transcriptomique and Génomique Marseille Luminy (TGML) Platform and supported by grants from INSERM, GIS Infrastructures en Biologie Santé et Agronomie (IBiSA), Aix-Marseille Université, and ANR-10-INBS-0009-10. This work was supported by La Ligue Contre le Cancer, Association pour la Recherche sur le Cancer (ARC), Institute National du Cancer (INCa), Cancéropôle Provence-Alpes-Côte d'Azur (PACA), and INSERM. Proteomic analyses were done using the mass spectrometry facility of Marseille Proteomics (
Figures

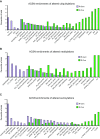

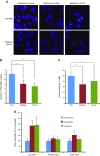



References
-
- Schneider G., Siveke J. T., Eckel F., Schmid R. M. (2005) Pancreatic cancer: basic and clinical aspects. Gastroenterology 128, 1606–1625 - PubMed
-
- Siegel R. L., Miller K. D., Jemal A. (2018) Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 - PubMed
-
- Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J. L., Gourgou-Bourgade S., de la Fouchardière C., Bennouna J., Bachet J. B., Khemissa-Akouz F., Péré-Vergé D., Delbaldo C., Assenat E., Chauffert B., Michel P., Montoto-Grillot C., Ducreux M.; Groupe Tumeurs Digestives of UnicancerPRODIGE Intergroup (2011) FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825 - PubMed
-
- Von Hoff D. D., Ervin T., Arena F. P., Chiorean E. G., Infante J., Moore M., Seay T., Tjulandin S. A., Ma W. W., Saleh M. N., Harris M., Reni M., Dowden S., Laheru D., Bahary N., Ramanathan R. K., Tabernero J., Hidalgo M., Goldstein D., Van Cutsem E., Wei X., Iglesias J., Renschler M. F. (2013) Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 369, 1691–1703 - PMC - PubMed
-
- Adamska A., Elaskalani O., Emmanouilidi A., Kim M., Abdol Razak N. B., Metharom P., Falasca M. (2018) Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv. Biol. Regul. 68, 77–87 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

