Ten simple rules to create biological network figures for communication
- PMID: 31557157
- PMCID: PMC6762067
- DOI: 10.1371/journal.pcbi.1007244
Ten simple rules to create biological network figures for communication
Abstract
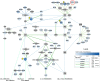
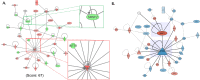
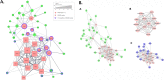
Biological network figures are ubiquitous in the biology and medical literature. On the one hand, a good network figure can quickly provide information about the nature and degree of interactions between items and enable inferences about the reason for those interactions. On the other hand, good network figures are difficult to create. In this paper, we outline 10 simple rules for creating biological network figures for communication, from choosing layouts, to applying color or other channels to show attributes, to the use of layering and separation. These rules are accompanied by illustrative examples. We also provide a concise set of references and additional resources for each rule.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

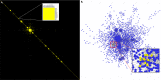

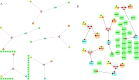
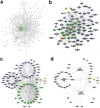
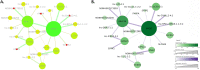

References
-
- Aerts J, Gehlenborg N, Marai GE, Nieselt KK. Visualization of Biological Data—Crossroads (Dagstuhl Seminar 18161). Dagstuhl Reports. 2018;8(4):32–71. 10.4230/DagRep.8.4.32 - DOI
-
- Nathalie Henry Riche; Christophe Hurter; Nicholas Diakopoulos; Sheelagh Carpendale. Data-Driven Storytelling. AK Peters; 2018.
-
- Munzner T. Visualization Analysis & Design A K Peters Visualization; CRC Press; 2014.

