Chirality-Dependent Adsorption between Amphipathic Peptide and POPC Membrane
- PMID: 31557910
- PMCID: PMC6801444
- DOI: 10.3390/ijms20194760
Chirality-Dependent Adsorption between Amphipathic Peptide and POPC Membrane
Abstract
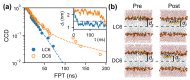
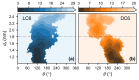
The interactions between chiral molecules and cell membranes have attracted more and more attention in recent decades, due to their importance in molecular science and medical applications. It is observed that some peptides composed of different chiral amino acids may have distinct interactions with a membrane. How does the membrane exhibit a selective behavior related to the chirality of the peptides? Microscopically, the interactions between the peptides and the membrane are poorly understood. In this work, we study the interactions between an amphipathic peptide (C6) and POPC membrane with simulations. The kinetics and thermodynamics of peptide enantiomers during the adsorption to the membrane are characterized with direct simulations and umbrella sampling. It is observed that there are slow kinetics for the peptide composed of D-type amino acids. Along the observed pathways, the free energy landscapes are determined with umbrella sampling techniques. A free-energy barrier for the peptide composed of D-amino acids is observed, which is consistent with the kinetic observations. The results indicate the concurrent adsorption and rotation of the peptide helix. The local interactions between the peptides and the membrane are examined in detail, including the contact interactions between the peptides and the membrane, and the distributions of the lipids around the peptide. There are observable differences of the local interactions for the cases related to different peptide enantiomers. These results further demonstrate the importance of the rotation of peptide helix during the adsorption. More interestingly, all these kinetic differences between peptide enantiomers can be explained based on the conformations of the residue Trp and interactions between Trp and lipid molecules. These results give us a molecular understanding of the mechanism of the chirality-dependent peptide-membrane interactions, and may provide clues to designing systems which are sensitive to the chirality of membranes.
Keywords: POPC; chirality; membrane; molecular dynamics; protein; tryptophan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Coupling molecular dynamics simulations with experiments for the rational design of indolicidin-analogous antimicrobial peptides.J Mol Biol. 2009 Sep 25;392(3):837-54. doi: 10.1016/j.jmb.2009.06.071. Epub 2009 Jul 2. J Mol Biol. 2009. PMID: 19576903
-
Molecular dynamics simulation of the membrane binding and disruption mechanisms by antimicrobial scorpion venom-derived peptides.J Biomol Struct Dyn. 2018 Jun;36(8):2070-2084. doi: 10.1080/07391102.2017.1341340. Epub 2017 Jun 22. J Biomol Struct Dyn. 2018. PMID: 28604248
-
Solid-state nuclear magnetic resonance relaxation studies of the interaction mechanism of antimicrobial peptides with phospholipid bilayer membranes.Biochemistry. 2005 Aug 2;44(30):10208-17. doi: 10.1021/bi050730p. Biochemistry. 2005. PMID: 16042398
-
Antimicrobial Peptide Simulations and the Influence of Force Field on the Free Energy for Pore Formation in Lipid Bilayers.J Chem Theory Comput. 2016 Sep 13;12(9):4524-33. doi: 10.1021/acs.jctc.6b00265. Epub 2016 Aug 30. J Chem Theory Comput. 2016. PMID: 27529120
-
Simulation studies of the interaction of antimicrobial peptides and lipid bilayers.Biochim Biophys Acta. 1999 Dec 15;1462(1-2):185-200. doi: 10.1016/s0005-2736(99)00206-0. Biochim Biophys Acta. 1999. PMID: 10590308 Review.
Cited by
-
Chirality Effects in Peptide Assembly Structures.Front Bioeng Biotechnol. 2021 Jun 22;9:703004. doi: 10.3389/fbioe.2021.703004. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34239866 Free PMC article. Review.
-
Quantitative Assessment of Chirality of Protein Secondary Structures and Phenylalanine Peptide Nanotubes.Nanomaterials (Basel). 2021 Dec 5;11(12):3299. doi: 10.3390/nano11123299. Nanomaterials (Basel). 2021. PMID: 34947648 Free PMC article.
-
Stereoselectivity of Interaction of Nonsteroidal Anti-Inflammatory Drug S-Ketoprofen with L/D-Tryptophan in Phospholipid Membranes.Membranes (Basel). 2022 Apr 24;12(5):460. doi: 10.3390/membranes12050460. Membranes (Basel). 2022. PMID: 35629787 Free PMC article.
-
Insight into the Mechanism of Action and Peptide-Membrane Interactions of Aib-Rich Peptides: Multitechnique Experimental and Theoretical Analysis.Chembiochem. 2021 May 4;22(9):1656-1667. doi: 10.1002/cbic.202000834. Epub 2021 Feb 24. Chembiochem. 2021. PMID: 33411956 Free PMC article.
References
-
- Savile C.K., Janey J.M., Mundorff E.C., Moore J.C., Tam S., Jarvis W.R., Colbeck J.C., Krebber A., Fleitz F.J., Brands J., et al. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

