Drosophila Heterochromatin Stabilization Requires the Zinc-Finger Protein Small Ovary
- PMID: 31558581
- PMCID: PMC6827387
- DOI: 10.1534/genetics.119.302590
Drosophila Heterochromatin Stabilization Requires the Zinc-Finger Protein Small Ovary
Abstract
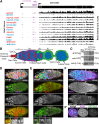
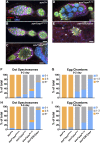
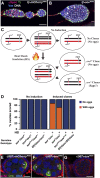
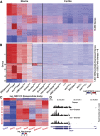
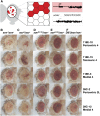
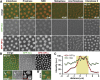
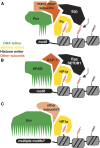
Heterochromatin-mediated repression is essential for controlling the expression of transposons and for coordinated cell type-specific gene regulation. The small ovary (sov) locus was identified in a screen for female-sterile mutations in Drosophila melanogaster, and mutants show dramatic ovarian morphogenesis defects. We show that the null sov phenotype is lethal and map the locus to the uncharacterized gene CG14438, which encodes a nuclear zinc-finger protein that colocalizes with the essential Heterochromatin Protein 1 (HP1a). We demonstrate Sov functions to repress inappropriate gene expression in the ovary, silence transposons, and suppress position-effect variegation in the eye, suggesting a central role in heterochromatin stabilization.
Keywords: HP1a; gene expression; heterochromatin; oogenesis; position-effect variegation; sov zinc-finger.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

