Mutations at hypothetical binding site 2 in insulin and insulin-like growth factors 1 and 2 result in receptor- and hormone-specific responses
- PMID: 31558604
- PMCID: PMC6873181
- DOI: 10.1074/jbc.RA119.010072
Mutations at hypothetical binding site 2 in insulin and insulin-like growth factors 1 and 2 result in receptor- and hormone-specific responses
Abstract
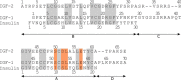
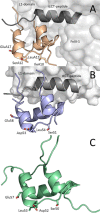
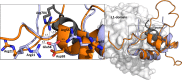
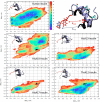
Information on how insulin and insulin-like growth factors 1 and 2 (IGF-1 and -2) activate insulin receptors (IR-A and -B) and the IGF-1 receptor (IGF-1R) is crucial for understanding the difference in the biological activities of these peptide hormones. Cryo-EM studies have revealed that insulin uses its binding sites 1 and 2 to interact with IR-A and have identified several critical residues in binding site 2. However, mutagenesis studies suggest that Ile-A10, Ser-A12, Leu-A13, and Glu-A17 also belong to insulin's site 2. Here, to resolve this discrepancy, we mutated these insulin residues and the equivalent residues in IGFs. Our findings revealed that equivalent mutations in the hormones can result in differential biological effects and that these effects can be receptor-specific. We noted that the insulin positions A10 and A17 are important for its binding to IR-A and IR-B and IGF-1R and that A13 is important only for IR-A and IR-B binding. The IGF-1/IGF-2 positions 51/50 and 54/53 did not appear to play critical roles in receptor binding, but mutations at IGF-1 position 58 and IGF-2 position 57 affected the binding. We propose that IGF-1 Glu-58 interacts with IGF-1R Arg-704 and belongs to IGF-1 site 1, a finding supported by the NMR structure of the less active Asp-58-IGF-1 variant. Computational analyses indicated that the aforementioned mutations can affect internal insulin dynamics and inhibit adoption of a receptor-bound conformation, important for binding to receptor site 1. We provide a molecular model and alternative hypotheses for how the mutated insulin residues affect activity.
Keywords: NMR structure; complex; hormone analog; insulin; insulin-like growth factor (IGF); molecular dynamics; mutagenesis; peptide hormone; receptor autophosphorylation; receptor binding; receptor tyrosine kinase; structural biology; structure-function.
© 2019 Macháčková et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

