How to copy and paste DNA microarrays
- PMID: 31558745
- PMCID: PMC6763488
- DOI: 10.1038/s41598-019-50371-1
How to copy and paste DNA microarrays
Abstract
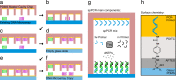
Analogous to a photocopier, we developed a DNA microarray copy technique and were able to copy patterned original DNA microarrays. With this process the appearance of the copied DNA microarray can also be altered compared to the original by producing copies of different resolutions. As a homage to the very first photocopy made by Chester Charlson and Otto Kornei, we performed a lookalike DNA microarray copy exactly 80 years later. Those copies were also used for label-free real-time kinetic binding assays of apo-dCas9 to double stranded DNA and of thrombin to single stranded DNA. Since each DNA microarray copy was made with only 5 µl of spPCR mix, the whole process is cost-efficient. Hence, our DNA microarray copier has a great potential for becoming a standard lab tool.
Conflict of interest statement
SDK, JW and GR are inventors of the presented method which is filed as pending patent (PCT/EP2019/062502).
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

