Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi
- PMID: 31559019
- PMCID: PMC6754632
- DOI: 10.1186/s40694-019-0076-7
Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi
Abstract
Background: CRISPR/Cas9 mediated genome editing has expedited the way of constructing multiple gene alterations in filamentous fungi, whereas traditional methods are time-consuming and can be of mutagenic nature. These developments allow the study of large gene families that contain putatively redundant genes, such as the seven-membered family of crh-genes encoding putative glucan-chitin crosslinking enzymes involved in cell wall biosynthesis.
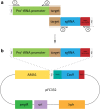
Results: Here, we present a CRISPR/Cas9 system for Aspergillus niger using a non-integrative plasmid, containing a selection marker, a Cas9 and a sgRNA expression cassette. Combined with selection marker free knockout repair DNA fragments, a set of the seven single knockout strains was obtained through homology directed repair (HDR) with an average efficiency of 90%. Cas9-sgRNA plasmids could effectively be cured by removing selection pressure, allowing the use of the same selection marker in successive transformations. Moreover, we show that either two or even three separate Cas9-sgRNA plasmids combined with marker-free knockout repair DNA fragments can be used in a single transformation to obtain double or triple knockouts with 89% and 38% efficiency, respectively. By employing this technique, a seven-membered crh-gene family knockout strain was acquired in a few rounds of transformation; three times faster than integrative selection marker (pyrG) recycling transformations. An additional advantage of the use of marker-free gene editing is that negative effects of selection marker gene expression are evaded, as we observed in the case of disrupting virtually silent crh family members.
Conclusions: Our findings advocate the use of CRISPR/Cas9 to create multiple gene deletions in both a fast and reliable way, while simultaneously omitting possible locus-dependent-side-effects of poor auxotrophic marker expression.
Keywords: Aspergillus niger; CRISPR/Cas9; Cell wall; Gene editing; Gene families; Knockouts; Marker free; Multiplexing; pyrG marker effects.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures



References
-
- Free SJ. Fungal cell wall organization and biosynthesis. 1. New York: Elsevier Inc.; 2013. - PubMed
LinkOut - more resources
Full Text Sources

